A membrane-bound NAC transcription factor regulates cell division in Arabidopsis
- PMID: 17098812
- PMCID: PMC1693948
- DOI: 10.1105/tpc.106.043018
A membrane-bound NAC transcription factor regulates cell division in Arabidopsis
Abstract
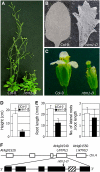
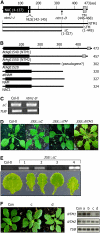
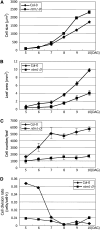
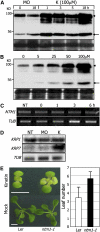
Controlled release of membrane-tethered, dormant precursors is an intriguing activation mechanism that regulates diverse cellular functions in eukaryotes. An exquisite example is the proteolytic activation of membrane-bound transcription factors. The proteolytic cleavage liberates active transcription factors from the membranes that can enter the nucleus and evokes rapid transcriptional responses to incoming stimuli. Here, we show that a membrane-bound NAC (for NAM, ATAF1/2, CUC2) transcription factor, designated NTM1 (for NAC with transmembrane motif1), is activated by proteolytic cleavage through regulated intramembrane proteolysis and mediates cytokinin signaling during cell division in Arabidopsis thaliana. Cell proliferation was greatly reduced in an Arabidopsis mutant with retarded growth and serrated leaves in which a transcriptionally active NTM1 form was constitutively expressed. Accordingly, a subset of cyclin-dependent kinase (CDK) inhibitor genes (the KIP-related proteins) was induced in this mutant with a significant reduction in histone H4 gene expression and in CDK activity. Consistent with a role for NTM1 in cell cycling, a Ds element insertional mutant was morphologically normal but displayed enhanced hypocotyl growth with accelerated cell division. Interestingly, cytokinins were found to regulate NTM1 activity by controlling its stability. These results indicate that the membrane-mediated activation of NTM1 defines a molecular mechanism by which cytokinin signaling is tightly regulated during cell cycling.
Figures



Similar articles
-
Membrane regulation of cytokinin-mediated cell division in Arabidopsis.Plant Signal Behav. 2007 Jan;2(1):15-6. doi: 10.4161/psb.2.1.3606. Plant Signal Behav. 2007. PMID: 19704799 Free PMC article.
-
Advances in membrane-tethered NAC transcription factors in plants.Plant Sci. 2024 May;342:112034. doi: 10.1016/j.plantsci.2024.112034. Epub 2024 Feb 15. Plant Sci. 2024. PMID: 38365003 Review.
-
A membrane-tethered transcription factor ANAC089 negatively regulates floral initiation in Arabidopsis thaliana.Sci China Life Sci. 2010 Nov;53(11):1299-306. doi: 10.1007/s11427-010-4085-2. Epub 2010 Nov 3. Sci China Life Sci. 2010. PMID: 21046321
-
Exploring membrane-associated NAC transcription factors in Arabidopsis: implications for membrane biology in genome regulation.Nucleic Acids Res. 2007;35(1):203-13. doi: 10.1093/nar/gkl1068. Epub 2006 Dec 7. Nucleic Acids Res. 2007. PMID: 17158162 Free PMC article.
-
Membrane-tethered transcription factors in Arabidopsis thaliana: novel regulators in stress response and development.Curr Opin Plant Biol. 2008 Dec;11(6):695-701. doi: 10.1016/j.pbi.2008.10.005. Curr Opin Plant Biol. 2008. PMID: 19019722 Review.
Cited by
-
RhNAC2 and RhEXPA4 are involved in the regulation of dehydration tolerance during the expansion of rose petals.Plant Physiol. 2012 Dec;160(4):2064-82. doi: 10.1104/pp.112.207720. Epub 2012 Oct 23. Plant Physiol. 2012. PMID: 23093360 Free PMC article.
-
Characterization of NAC transcription factor NtNAC028 as a regulator of leaf senescence and stress responses.Front Plant Sci. 2022 Aug 15;13:941026. doi: 10.3389/fpls.2022.941026. eCollection 2022. Front Plant Sci. 2022. PMID: 36046590 Free PMC article.
-
Functional Characterization of the CpNAC1 Promoter and Gene from Chimonanthus praecox in Arabidopsis.Int J Mol Sci. 2022 Dec 29;24(1):542. doi: 10.3390/ijms24010542. Int J Mol Sci. 2022. PMID: 36613984 Free PMC article.
-
A membrane-associated NAC transcription factor regulates salt-responsive flowering via FLOWERING LOCUS T in Arabidopsis.Planta. 2007 Aug;226(3):647-54. doi: 10.1007/s00425-007-0513-3. Epub 2007 Apr 5. Planta. 2007. PMID: 17410378
-
OoNAC72, a NAC-Type Oxytropis ochrocephala Transcription Factor, Conferring Enhanced Drought and Salt Stress Tolerance in Arabidopsis.Front Plant Sci. 2019 Jul 11;10:890. doi: 10.3389/fpls.2019.00890. eCollection 2019. Front Plant Sci. 2019. PMID: 31354764 Free PMC article.
References
-
- Ahn, J., Kim, M., Lim, J.H., Kim, K.-T., and Pai, H.-S. (2004). Phytocalpain controls the proliferation and differentiation fates of cells in plant organ development. Plant J. 38 969–981. - PubMed
-
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815. - PubMed
-
- Brown, M.S., Ye, J., Rawson, R.B., and Goldstein, J.L. (2000). Regulated intramembrane proteolysis: A control mechanism conserved from bacteria to humans. Cell 100 391–398. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

