Ectopic expression of a basic helix-loop-helix gene transactivates parallel pathways of proanthocyanidin biosynthesis. structure, expression analysis, and genetic control of leucoanthocyanidin 4-reductase and anthocyanidin reductase genes in Lotus corniculatus
- PMID: 17098849
- PMCID: PMC1761954
- DOI: 10.1104/pp.106.090886
Ectopic expression of a basic helix-loop-helix gene transactivates parallel pathways of proanthocyanidin biosynthesis. structure, expression analysis, and genetic control of leucoanthocyanidin 4-reductase and anthocyanidin reductase genes in Lotus corniculatus
Abstract
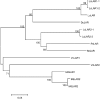
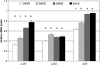
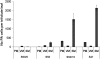
Proanthocyanidins (PAs) are plant secondary metabolites and are composed primarily of catechin and epicatechin units in higher plant species. Due to the ability of PAs to bind reversibly with plant proteins to improve digestion and reduce bloat, engineering this pathway in leaves is a major goal for forage breeders. Here, we report the cloning and expression analysis of anthocyanidin reductase (ANR) and leucoanthocyanidin 4-reductase (LAR), two genes encoding enzymes committed to epicatechin and catechin biosynthesis, respectively, in Lotus corniculatus. We show the presence of two LAR gene families (LAR1 and LAR2) and that the steady-state levels of ANR and LAR1 genes correlate with the levels of PAs in leaves of wild-type and transgenic plants. Interestingly, ANR and LAR1, but not LAR2, genes produced active proteins following heterologous expression in Escherichia coli and are affected by the same basic helix-loop-helix transcription factor that promotes PA accumulation in cells of palisade and spongy mesophyll. This study provides direct evidence that the same subclass of transcription factors can mediate the expression of the structural genes of both branches of PA biosynthesis.
Figures





References
-
- Abrahams S, Lee E, Walker AR, Tanner GJ, Larkin PJ, Ashton AR (2003) The Arabidopsis TDS4 gene encodes leucoanthocyanidin dioxygenase (LDOX) and is essential for proanthocyanidin synthesis and vacuole development. Plant J 35 624–636 - PubMed
-
- Barry TN, McNabb WC (1999) The implications of condensed tannins on the nutritive value of temperate forages fed to ruminants. Br J Nutr 81 263–272 - PubMed
-
- Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L (2004) TT2, TT8 and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J 39 366–380 - PubMed
-
- Baxter IR, Young JC, Armstrong G, Foster N, Bogenschutz N, Cordova T, Peer WA, Hazen SP, Murphy AS, Harper JF (2005) A plasma membrane H+-ATPase is required for the formation of proanthocyanidins in the seed coat endothelium of Arabidopsis thaliana. Proc Natl Acad Sci USA 102: 2649–2654; erratum Baxter IR, Young JC, Armstrong G, Foster N, Bogenschutz N, Cordova T, Peer WA, Hazen SP, Murphy AS, Harper JF (2005) Proc Natl Acad Sci USA 102: 5635 - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

