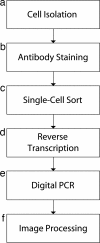
Transcription factor profiling in individual hematopoietic progenitors by digital RT-PCR
- PMID: 17098862
- PMCID: PMC1693828
- DOI: 10.1073/pnas.0608512103
Transcription factor profiling in individual hematopoietic progenitors by digital RT-PCR
Abstract
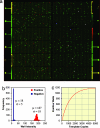
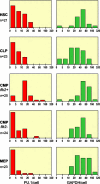
We report here a systematic, quantitative population analysis of transcription factor expression within developmental progenitors, made possible by a microfluidic chip-based "digital RT-PCR" assay that can count template molecules in cDNA samples prepared from single cells. In a survey encompassing five classes of early hematopoietic precursor, we found markedly heterogeneous expression of the transcription factor PU.1 in hematopoietic stem cells and divergent patterns of PU.1 expression within flk2- and flk2+ common myeloid progenitors. The survey also revealed significant differences in the level of the housekeeping transcript GAPDH across the surveyed populations, which demonstrates caveats of normalizing expression data to endogenous controls and underscores the need to put gene measurement on an absolute, copy-per-cell basis.
Conflict of interest statement
Conflict of interest statement: S.Q. is a founder, shareholder, and consultant for Fluidigm Corporation.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

