Functional analysis of luxS in the probiotic strain Lactobacillus rhamnosus GG reveals a central metabolic role important for growth and biofilm formation
- PMID: 17098890
- PMCID: PMC1797292
- DOI: 10.1128/JB.01394-06
Functional analysis of luxS in the probiotic strain Lactobacillus rhamnosus GG reveals a central metabolic role important for growth and biofilm formation
Abstract
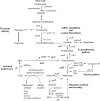
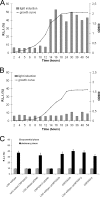
Quorum sensing is involved in the regulation of multicellular behavior through communication via small molecules. Given the high number and diversity of the gastrointestinal microbiota, it is postulated that members of this community communicate to coordinate a variety of adaptive processes. AI-2 is suggested to be a universal bacterial signaling molecule synthesized by the LuxS enzyme, which forms an integral part of the activated methyl cycle. We have previously reported that the well-documented probiotic strain Lactobacillus rhamnosus GG, a human isolate, produces AI-2-like molecules. In this study, we identified the luxS homologue of L. rhamnosus GG. luxS seems to be located in an operon with a yxjH gene encoding a putative cobalamin-independent methionine synthase. In silico analysis revealed a methionine-specific T box in the leader sequence of the putative yxjH-luxS operon. However, transcriptional analysis showed that luxS is expressed mainly as a monocistronic transcript. Construction of a luxS knockout mutant confirmed that the luxS gene is responsible for AI-2 production in L. rhamnosus GG. However, this mutation also resulted in pleiotropic effects on the growth of this fastidious strain. Cysteine, pantothenate, folic acid, and biotin could partially complement growth, suggesting a central metabolic role for luxS in L. rhamnosus GG. Interestingly, the luxS mutant also showed a defect in monospecies biofilm formation. Experiments with chemically synthesized (S)-4,5-dihydroxy-2,3-pentanedione, coculture with the wild type, and nutritional complementation suggested that the main cause of this defect has a metabolic nature. Moreover, our data indicate that suppressor mutations are likely to occur in luxS mutants of L. rhamnosus GG. Therefore, results of luxS-related studies should be carefully interpreted.
Figures

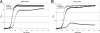
References
-
- Altermann, E., W. M. Russell, M. A. Azcarate-Peril, R. Barrangou, B. L. Buck, O. McAuliffe, N. Souther, A. Dobson, T. Duong, M. Callanan, S. Lick, A. Hamrick, R. Cano, and T. R. Klaenhammer. 2005. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. USA 102:3906-3912. - PMC - PubMed
-
- Alvarez, M. A., M. Herrero, and J. E. Suarez. 1998. The site-specific recombination system of the Lactobacillus species bacteriophage A2 integrates in gram-positive and gram-negative bacteria. Virology 250:185-193. - PubMed
-
- Bassler, B. L., M. Wright, R. E. Showalter, and M. R. Silverman. 1993. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol. Microbiol. 9:773-786. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

