Analysis of the amino acid sequence specificity determinants of the enterococcal cCF10 sex pheromone in interactions with the pheromone-sensing machinery
- PMID: 17098891
- PMCID: PMC1797347
- DOI: 10.1128/JB.01226-06
Analysis of the amino acid sequence specificity determinants of the enterococcal cCF10 sex pheromone in interactions with the pheromone-sensing machinery
Abstract
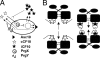
The level of expression of conjugation genes in Enterococcus faecalis strains carrying the pheromone-responsive transferable plasmid pCF10 is determined by the ratio in the culture medium of two types of signaling peptides, a pheromone (cCF10) and an inhibitor (iCF10). Recent data have demonstrated that both peptides target the cytoplasmic receptor protein PrgX. However, the relative importance of the interaction of these peptides with the pCF10 protein PrgZ (which enhances import of cCF10) versus PrgX is not fully understood, and there is relatively little information about specific amino acid sequence determinants affecting the functional interactions of cCF10 with these proteins in vivo. To address these issues, we used a pheromone-inducible reporter gene system where various combinations of PrgX and PrgZ could be expressed in an isogenic host background to examine the biological activities of cCF10, iCF10, and variants of cCF10 isolated in a genetic screen. The results suggest that most of the amino acid sequence determinants of cCF10 pheromone activity affect interactions between the peptide and PrgX, although some sequence variants that affected peptide/PrgZ interactions were also identified. The results provide functional data to complement ongoing structural studies of PrgX and increase our understanding of the functional interactions of cCF10 and iCF10 with the pheromone-sensing machinery of pCF10.
Figures



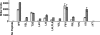
Similar articles
-
Characterization of the sequence specificity determinants required for processing and control of sex pheromone by the intramembrane protease Eep and the plasmid-encoded protein PrgY.J Bacteriol. 2008 Feb;190(4):1172-83. doi: 10.1128/JB.01327-07. Epub 2007 Dec 14. J Bacteriol. 2008. PMID: 18083822 Free PMC article.
-
Enterococcus faecalis pheromone-responsive protein PrgX: genetic separation of positive autoregulatory functions from those involved in negative regulation of conjugative plasmid transfer.Mol Microbiol. 2004 Oct;54(2):520-32. doi: 10.1111/j.1365-2958.2004.04286.x. Mol Microbiol. 2004. PMID: 15469521
-
Cloning and characterization of a region of the Enterococcus faecalis conjugative plasmid, pCF10, encoding a sex pheromone-binding function.J Bacteriol. 1993 Aug;175(16):5253-9. doi: 10.1128/jb.175.16.5253-5259.1993. J Bacteriol. 1993. PMID: 8349565 Free PMC article.
-
Enterococcal Sex Pheromones: Evolutionary Pathways to Complex, Two-Signal Systems.J Bacteriol. 2016 May 13;198(11):1556-1562. doi: 10.1128/JB.00128-16. Print 2016 Jun 1. J Bacteriol. 2016. PMID: 27021562 Free PMC article. Review.
-
The peptide pheromone-inducible conjugation system of Enterococcus faecalis plasmid pCF10: cell-cell signalling, gene transfer, complexity and evolution.Philos Trans R Soc Lond B Biol Sci. 2007 Jul 29;362(1483):1185-93. doi: 10.1098/rstb.2007.2043. Philos Trans R Soc Lond B Biol Sci. 2007. PMID: 17360276 Free PMC article. Review.
Cited by
-
Structural Differences in Complexes between the Master Regulator PrgX, Peptide Pheromones, and Operator Binding Sites Determine the Induction State for Conjugative Transfer of pCF10.J Bacteriol. 2022 Dec 20;204(12):e0029822. doi: 10.1128/jb.00298-22. Epub 2022 Nov 10. J Bacteriol. 2022. PMID: 36354318 Free PMC article.
-
Characterization of the sequence specificity determinants required for processing and control of sex pheromone by the intramembrane protease Eep and the plasmid-encoded protein PrgY.J Bacteriol. 2008 Feb;190(4):1172-83. doi: 10.1128/JB.01327-07. Epub 2007 Dec 14. J Bacteriol. 2008. PMID: 18083822 Free PMC article.
-
Effects of endogenous levels of master regulator PrgX and peptide pheromones on inducibility of conjugation in the enterococcal pCF10 system.Mol Microbiol. 2019 Sep;112(3):1010-1023. doi: 10.1111/mmi.14339. Epub 2019 Jul 18. Mol Microbiol. 2019. PMID: 31265752 Free PMC article.
-
Structural analysis of the Anti-Q-Qs interaction: RNA-mediated regulation of E. faecalis plasmid pCF10 conjugation.Plasmid. 2010 Jul;64(1):26-35. doi: 10.1016/j.plasmid.2010.03.002. Epub 2010 Mar 21. Plasmid. 2010. PMID: 20332003 Free PMC article.
-
Antagonistic self-sensing and mate-sensing signaling controls antibiotic-resistance transfer.Proc Natl Acad Sci U S A. 2013 Apr 23;110(17):7086-90. doi: 10.1073/pnas.1212256110. Epub 2013 Apr 8. Proc Natl Acad Sci U S A. 2013. PMID: 23569272 Free PMC article.
References
-
- An, F. Y., and D. B. Clewell. 1994. Characterization of the determinant (traB) encoding sex pheromone shutdown by the hemolysin/bacteriocin plasmid, pAD1 in Enterococcus faecalis. Plasmid 31:215-221. - PubMed
-
- Bae, T., S. Clerc-Bardin, and G. M. Dunny. 2000. Analysis of expression of prgX, a key negative regulator of the transfer of the Enterococcus faecalis pheromone-inducible plasmid pCF10. J. Mol. Biol. 297:861-875. - PubMed
-
- Bae, T., and G. M. Dunny. 2001. Dominant negative mutants of prgX: evidence for a role of PrgX dimerization in negative regulation of pheromone-inducible conjugation. Mol. Microbiol. 39:1307-1320. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

