The complete genome sequence of Roseobacter denitrificans reveals a mixotrophic rather than photosynthetic metabolism
- PMID: 17098896
- PMCID: PMC1797316
- DOI: 10.1128/JB.01390-06
The complete genome sequence of Roseobacter denitrificans reveals a mixotrophic rather than photosynthetic metabolism
Abstract
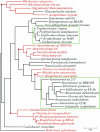
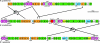
Purple aerobic anoxygenic phototrophs (AAPs) are the only organisms known to capture light energy to enhance growth only in the presence of oxygen but do not produce oxygen. The highly adaptive AAPs compose more than 10% of the microbial community in some euphotic upper ocean waters and are potentially major contributors to the fixation of the greenhouse gas CO2. We present the complete genomic sequence and feature analysis of the AAP Roseobacter denitrificans, which reveal clues to its physiology. The genome lacks genes that code for known photosynthetic carbon fixation pathways, and most notably missing are genes for the Calvin cycle enzymes ribulose bisphosphate carboxylase (RuBisCO) and phosphoribulokinase. Phylogenetic evidence implies that this absence could be due to a gene loss from a RuBisCO-containing alpha-proteobacterial ancestor. We describe the potential importance of mixotrophic rather than autotrophic CO2 fixation pathways in these organisms and suggest that these pathways function to fix CO2 for the formation of cellular components but do not permit autotrophic growth. While some genes that code for the redox-dependent regulation of photosynthetic machinery are present, many light sensors and transcriptional regulatory motifs found in purple photosynthetic bacteria are absent.
Figures


References
-
- Ashida, H., A. Danchin, and A. Yokota. 2005. Was photosynthetic RuBisCO recruited by acquisitive evolution from RuBisCO-like proteins involved in sulfur metabolism? Res. Microbiol. 156:611-618. - PubMed
-
- Bauer, C. E. 2004. Regulation of photosystem synthesis in Rhodobacter capsulatus. Photosynth. Res. 80:353-360. - PubMed
-
- Beatty, J. T. 1995. Organization of photosynthesis gene transcripts, p. 1209-1219. In R. E. Blankenship, M. T. Madigan, and C. E. Bauer (ed.), Anoxygenic photosynthetic bacteria, vol. 2. Kluwer Academic Publishers, Dordrecht, The Netherlands.
-
- Braatsch, S., M. Gomelsky, S. Kuphal, and G. Klug. 2002. A single flavoprotein, AppA, integrates both redox and light signals in Rhodobacter sphaeroides. Mol. Microbiol. 45:827-836. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

