NCBI GEO: mining tens of millions of expression profiles--database and tools update
- PMID: 17099226
- PMCID: PMC1669752
- DOI: 10.1093/nar/gkl887
NCBI GEO: mining tens of millions of expression profiles--database and tools update
Abstract
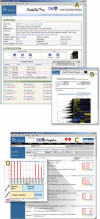
The Gene Expression Omnibus (GEO) repository at the National Center for Biotechnology Information (NCBI) archives and freely disseminates microarray and other forms of high-throughput data generated by the scientific community. The database has a minimum information about a microarray experiment (MIAME)-compliant infrastructure that captures fully annotated raw and processed data. Several data deposit options and formats are supported, including web forms, spreadsheets, XML and Simple Omnibus Format in Text (SOFT). In addition to data storage, a collection of user-friendly web-based interfaces and applications are available to help users effectively explore, visualize and download the thousands of experiments and tens of millions of gene expression patterns stored in GEO. This paper provides a summary of the GEO database structure and user facilities, and describes recent enhancements to database design, performance, submission format options, data query and retrieval utilities. GEO is accessible at http://www.ncbi.nlm.nih.gov/geo/
Figures
References
-
- Brazma A., Hingamp P., Quackenbush J., Sherlock G., Spellman P., Stoeckert C., Aach J., Ansorge W., Ball C.A., Causton H.C., et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nature Genet. 2001;29:365–371. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

