FlyBase: genomes by the dozen
- PMID: 17099233
- PMCID: PMC1669768
- DOI: 10.1093/nar/gkl827
FlyBase: genomes by the dozen
Abstract
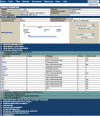
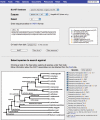
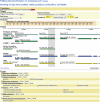
FlyBase (http://flybase.org/) is the primary database of genetic and genomic data for the insect family Drosophilidae. Historically, Drosophila melanogaster has been the most extensively studied species in this family, but recent determination of the genomic sequences of an additional 11 Drosophila species opens up new avenues of research for other Drosophila species. This extensive sequence resource, encompassing species with well-defined phylogenetic relationships, provides a model system for comparative genomic analyses. FlyBase has developed tools to facilitate access to and navigation through this invaluable new data collection.
Figures

References
-
- Zhou P., Emmert D., Zhang P. Using chado to store genome annotation data. In: Baxevanis A.D., Davison D.B., editors. Current Protocols in Bioinformatics. Vol. 2. Hoboken, NJ: John Wiley & Sons, Inc.; 2005. pp. 9.6.1–9.6.28. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

