Emergence of primate genes by retrotransposon-mediated sequence transduction
- PMID: 17101974
- PMCID: PMC1693794
- DOI: 10.1073/pnas.0603224103
Emergence of primate genes by retrotransposon-mediated sequence transduction
Abstract
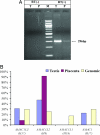
Gene duplication is one of the most important mechanisms for creating new genes and generating genomic novelty. Retrotransposon-mediated sequence transduction (i.e., the process by which a retrotransposon carries flanking sequence during its mobilization) has been proposed as a gene duplication mechanism. L1 exon shuffling potential has been reported in cell culture assays, and two potential L1-mediated exon shuffling events have been identified in the genome. SVA is the youngest retrotransposon family in primates and is capable of 3' flanking sequence transduction during retrotransposition. In this study, we examined all of the full-length SVA elements in the human genome to assess the frequency and impact of SVA-mediated 3' sequence transduction. Our results showed that approximately 53 kb of genomic sequences have been duplicated by 143 different SVA-mediated transduction events. In particular, we identified one group of SVA elements that duplicated the entire AMAC gene three times in the human genome through SVA-mediated transduction events, which happened before the divergence of humans and African great apes. In addition to the original AMAC gene, the three transduced AMAC copies contain intact ORFs in the human genome, and at least two are actively transcribed in different human tissues. The duplication of entire genes and the creation of previously undescribed gene families through retrotransposon-mediated sequence transduction represent an important mechanism by which mobile elements impact their host genomes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

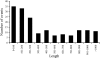


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

