Evolution and expression of chimeric POTE-actin genes in the human genome
- PMID: 17101985
- PMCID: PMC1693842
- DOI: 10.1073/pnas.0608344103
Evolution and expression of chimeric POTE-actin genes in the human genome
Abstract
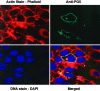
We previously described a primate-specific gene family, POTE, that is expressed in many cancers but in a limited number of normal organs. The 13 POTE genes are dispersed among eight different chromosomes and evolved by duplications and remodeling of the human genome from an ancestral gene, ANKRD26. Based on sequence similarity, the POTE gene family members can be divided into three groups. By genome database searches, we identified an actin retroposon insertion at the carboxyl terminus of one of the ancestral POTE paralogs. By Northern blot analysis, we identified the expected 7.5-kb POTE-actin chimeric transcript in a breast cancer cell line. The protein encoded by the POTE-actin transcript is predicted to be 120 kDa in size. Using anti-POTE mAbs that recognize the amino-terminal portion of the POTE protein, we detected the 120-kDa POTE-actin fusion protein in breast cancer cell lines known to express the fusion transcript. These data demonstrate that insertion of a retroposon produced an altered functional POTE gene. This example indicates that new functional human genes can evolve by insertion of retroposons.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Bera TK, Huynh N, Maeda H, Sathyanarayana BK, Lee B, Pastan I. Gene. 2004;337:45–53. - PubMed
-
- Hahn Y, Bera TK, Pastan IH, Lee B. Gene. 2006;366:238–245. - PubMed
-
- Bera TK, Saint-Fleur A, Lee Y, Kydd A, Hahn Y, Popescu NC, Zimonjic DB, Lee BK, Pastan I. Cancer Res. 2006;66:52–56. - PubMed
-
- Long M, Betrán E, Thornton K, Wang W. Nat Rev Genet. 2003;4:865–875. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

