Biological consequences of tightly bent DNA: the other life of a macromolecular celebrity
- PMID: 17103419
- PMCID: PMC3496788
- DOI: 10.1002/bip.20627
Biological consequences of tightly bent DNA: the other life of a macromolecular celebrity
Abstract
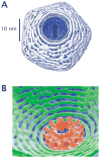
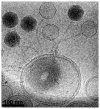
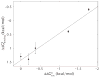
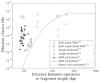
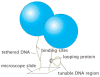
The mechanical properties of DNA play a critical role in many biological functions. For example, DNA packing in viruses involves confining the viral genome in a volume (the viral capsid) with dimensions that are comparable to the DNA persistence length. Similarly, eukaryotic DNA is packed in DNA-protein complexes (nucleosomes), in which DNA is tightly bent around protein spools. DNA is also tightly bent by many proteins that regulate transcription, resulting in a variation in gene expression that is amenable to quantitative analysis. In these cases, DNA loops are formed with lengths that are comparable to or smaller than the DNA persistence length. The aim of this review is to describe the physical forces associated with tightly bent DNA in all of these settings and to explore the biological consequences of such bending, as increasingly accessible by single-molecule techniques.
(c) 2006 Wiley Periodicals, Inc.
Figures

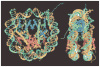


References
-
- Boal DH. Mechanics of the Cell. Cambridge University Press; Cambridge, England: 2002.
-
- Nelson P. Biological Physics: Energy, Information, Life. Freeman; New York: 2004.
-
- McCauley MJ, Williams MC. 2007;85:154–168. - PubMed
-
- Wiggins PA, Heijde Tvd, Moreno-Herrero F, Spakowitz A, Phillips R, Widom J, Dekker C, Nelson PC. Nat Nanotechnol. 2006;1:137–141. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

