The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure
- PMID: 17105344
- PMCID: PMC1618416
- DOI: 10.1371/journal.pbio.0040362
The liganding of glycolipid transfer protein is controlled by glycolipid acyl structure
Abstract
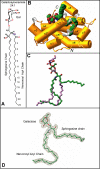
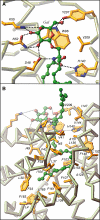
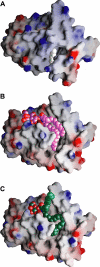
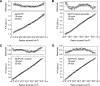
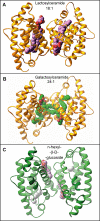
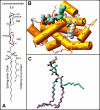
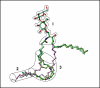
Glycosphingolipids (GSLs) play major roles in cellular growth and development. Mammalian glycolipid transfer proteins (GLTPs) are potential regulators of cell processes mediated by GSLs and display a unique architecture among lipid binding/transfer proteins. The GLTP fold represents a novel membrane targeting/interaction domain among peripheral proteins. Here we report crystal structures of human GLTP bound to GSLs of diverse acyl chain length, unsaturation, and sugar composition. Structural comparisons show a highly conserved anchoring of galactosyl- and lactosyl-amide headgroups by the GLTP recognition center. By contrast, acyl chain chemical structure and occupancy of the hydrophobic tunnel dictate partitioning between sphingosine-in and newly-observed sphingosine-out ligand-binding modes. The structural insights, combined with computed interaction propensity distributions, suggest a concerted sequence of events mediated by GLTP conformational changes during GSL transfer to and/or from membranes, as well as during GSL presentation and/or transfer to other proteins.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures



Comment in
-
Too long, too short, or just right: glycosphingolipid-protein binding varies with acyl chain length.PLoS Biol. 2006 Nov;4(11):e397. doi: 10.1371/journal.pbio.0040397. Epub 2006 Oct 24. PLoS Biol. 2006. PMID: 20076493 Free PMC article. No abstract available.
References
-
- Dwek RA, Butters TD, Platt FM, Zitzmann N. Targeting glycosylation as a therapeutic approach. Nat Rev Drug Discov. 2002;1:65–75. - PubMed
-
- Kozireski-Chuback D, Wu G, Ledeen RW. Upregulation of nuclear GM1 accompanies axon-like, but not dendrite-like, outgrowth in NG108–15 cells. J Neurosci Res. 1999;55:107–118. - PubMed
-
- Rippo MR, Malisan F, Ravagnan L, Tomassini B, Condo I, et al. GD3 ganglioside directly targets mitochondria in a bcl-2-controlled fashion. FASEB J. 2000;14:2047–2054. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

