Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the ESAT-6 (esx) gene cluster regions
- PMID: 17105670
- PMCID: PMC1660551
- DOI: 10.1186/1471-2148-6-95
Evolution and expansion of the Mycobacterium tuberculosis PE and PPE multigene families and their association with the duplication of the ESAT-6 (esx) gene cluster regions
Abstract
Background: The PE and PPE multigene families of Mycobacterium tuberculosis comprise about 10% of the coding potential of the genome. The function of the proteins encoded by these large gene families remains unknown, although they have been proposed to be involved in antigenic variation and disease pathogenesis. Interestingly, some members of the PE and PPE families are associated with the ESAT-6 (esx) gene cluster regions, which are regions of immunopathogenic importance, and encode a system dedicated to the secretion of members of the potent T-cell antigen ESAT-6 family. This study investigates the duplication characteristics of the PE and PPE gene families and their association with the ESAT-6 gene clusters, using a combination of phylogenetic analyses, DNA hybridization, and comparative genomics, in order to gain insight into their evolutionary history and distribution in the genus Mycobacterium.
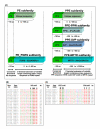
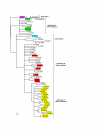
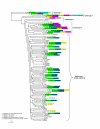
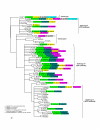
Results: The results showed that the expansion of the PE and PPE gene families is linked to the duplications of the ESAT-6 gene clusters, and that members situated in and associated with the clusters represent the most ancestral copies of the two gene families. Furthermore, the emergence of the repeat protein PGRS and MPTR subfamilies is a recent evolutionary event, occurring at defined branching points in the evolution of the genus Mycobacterium. These gene subfamilies are thus present in multiple copies only in the members of the M. tuberculosis complex and close relatives. The study provides a complete analysis of all the PE and PPE genes found in the sequenced genomes of members of the genus Mycobacterium such as M. smegmatis, M. avium paratuberculosis, M. leprae, M. ulcerans, and M. tuberculosis.
Conclusion: This work provides insight into the evolutionary history for the PE and PPE gene families of the mycobacteria, linking the expansion of these families to the duplications of the ESAT-6 (esx) gene cluster regions, and showing that they are composed of subgroups with distinct evolutionary (and possibly functional) differences.
Figures






References
-
- Hsu T, Hingley-Wilson SM, Chen B, Chen M, Dai AZ, Morin PM, Marks CB, Padiyar J, Goulding C, Gingery M, Eisenberg D, Russell RG, Derrick SC, Collins FM, Morris SL, King CH, Jacobs WR., Jr. The primary mechanism of attenuation of bacillus Calmette-Guerin is a loss of secreted lytic function required for invasion of lung interstitial tissue. Proc Natl Acad Sci U S A. 2003;100:12420–12425. doi: 10.1073/pnas.1635213100. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

