The CRM domain: an RNA binding module derived from an ancient ribosome-associated protein
- PMID: 17105995
- PMCID: PMC1705760
- DOI: 10.1261/rna.139607
The CRM domain: an RNA binding module derived from an ancient ribosome-associated protein
Abstract
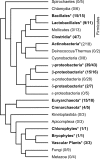
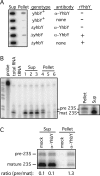
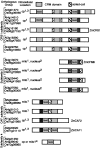
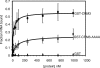
The CRS1-YhbY domain (also called the CRM domain) is represented as a stand-alone protein in Archaea and Bacteria, and in a family of single- and multidomain proteins in plants. The function of this domain is unknown, but structural data and the presence of the domain in several proteins known to interact with RNA have led to the proposal that it binds RNA. Here we describe a phylogenetic analysis of the domain, its incorporation into diverse proteins in plants, and biochemical properties of a prokaryotic and eukaryotic representative of the domain family. We show that a bacterial member of the family, Escherichia coli YhbY, is associated with pre-50S ribosomal subunits, suggesting that YhbY functions in ribosome assembly. GFP fused to a single-domain CRM protein from maize localizes to the nucleolus, suggesting that an analogous activity may have been retained in plants. We show further that an isolated maize CRM domain has RNA binding activity in vitro, and that a small motif shared with KH RNA binding domains, a conserved "GxxG" loop, contributes to its RNA binding activity. These and other results suggest that the CRM domain evolved in the context of ribosome function prior to the divergence of Archaea and Bacteria, that this function has been maintained in extant prokaryotes, and that the domain was recruited to serve as an RNA binding module during the evolution of plant genomes.
Figures




References
-
- Ban, N., Nissen, P., Hansen, J., Moore, P., Steitz, T. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. - PubMed
-
- Bremer, H., Dennis, P. Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt F.C., editor. Escherichia coli and Salmonella: Cellular and molecular biology. Vol. 2. ASM Press; Washington, DC: 1996. pp. 1553–1569.
-
- Bugl, H., Fauman, E., Staker, B., Zheng, F., Kushner, S., Saper, M., Bardwell, J., Jakob, U. RNA methylation under heat shock control. Mol. Cell. 2000;6:349–360. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
