Microsatellite signature of ecological selection for salt tolerance in a wild sunflower hybrid species, Helianthus paradoxus
- PMID: 17107488
- PMCID: PMC2442927
- DOI: 10.1111/j.1365-294X.2006.03112.x
Microsatellite signature of ecological selection for salt tolerance in a wild sunflower hybrid species, Helianthus paradoxus
Abstract
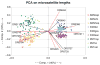
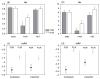
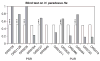
The hybrid sunflower species Helianthus paradoxus inhabits sporadic salt marshes in New Mexico and southwest Texas, USA, whereas its parental species, Helianthus annuus and Helianthus petiolaris, are salt sensitive. Previous studies identified three genomic regions - survivorship quantitative trait loci (QTLs) - that were under strong selection in experimental hybrids transplanted into the natural habitat of H. paradoxus. Here we ask whether these same genomic regions experienced significant selection during the origin and evolution of the natural hybrid, H. paradoxus. This was accomplished by comparing the variability of microsatellites linked to the three survivorship QTLs with those from genomic regions that were neutral in the experimental hybrids. As predicted if one or more selective sweeps had occurred in these regions, microsatellites linked to the survivorship QTLs exhibited a significant reduction in diversity in populations of the natural hybrid species. In contrast, no difference in diversity levels was observed between the two microsatellite classes in parental populations.
Figures
Similar articles
-
Natural selection for salt tolerance quantitative trait loci (QTLs) in wild sunflower hybrids: implications for the origin of Helianthus paradoxus, a diploid hybrid species.Mol Ecol. 2003 May;12(5):1225-35. doi: 10.1046/j.1365-294x.2003.01803.x. Mol Ecol. 2003. PMID: 12694286
-
The origin of ecological divergence in Helianthus paradoxus (Asteraceae): selection on transgressive characters in a novel hybrid habitat.Evolution. 2003 Sep;57(9):1989-2000. doi: 10.1111/j.0014-3820.2003.tb00379.x. Evolution. 2003. PMID: 14575321
-
Patterns of genetic diversity and candidate genes for ecological divergence in a homoploid hybrid sunflower, Helianthus anomalus.Mol Ecol. 2007 Dec;16(23):5017-29. doi: 10.1111/j.1365-294X.2007.03557.x. Epub 2007 Oct 16. Mol Ecol. 2007. PMID: 17944850 Free PMC article.
-
Response to salinity in the homoploid hybrid species Helianthus paradoxus and its progenitors H. annuus and H. petiolaris.New Phytol. 2006;170(3):615-29. doi: 10.1111/j.1469-8137.2006.01687.x. New Phytol. 2006. PMID: 16626481 Free PMC article.
-
The ecological genetics of homoploid hybrid speciation.J Hered. 2005 May-Jun;96(3):241-52. doi: 10.1093/jhered/esi026. Epub 2004 Dec 23. J Hered. 2005. PMID: 15618301 Free PMC article. Review.
Cited by
-
Genetic diversity and variation of Chinese fir from Fujian province and Taiwan, China, based on ISSR markers.PLoS One. 2017 Apr 13;12(4):e0175571. doi: 10.1371/journal.pone.0175571. eCollection 2017. PLoS One. 2017. PMID: 28406956 Free PMC article.
-
Applications of Molecular Markers for Developing Abiotic-Stress-Resilient Oilseed Crops.Life (Basel). 2022 Dec 28;13(1):88. doi: 10.3390/life13010088. Life (Basel). 2022. PMID: 36676037 Free PMC article. Review.
-
Natural selection and the genetics of adaptation in threespine stickleback.Philos Trans R Soc Lond B Biol Sci. 2010 Aug 27;365(1552):2479-86. doi: 10.1098/rstb.2010.0036. Philos Trans R Soc Lond B Biol Sci. 2010. PMID: 20643737 Free PMC article.
-
A coastal cline in sodium accumulation in Arabidopsis thaliana is driven by natural variation of the sodium transporter AtHKT1;1.PLoS Genet. 2010 Nov 11;6(11):e1001193. doi: 10.1371/journal.pgen.1001193. PLoS Genet. 2010. PMID: 21085628 Free PMC article.
-
Hitchhiking both ways: effect of two interfering selective sweeps on linked neutral variation.Genetics. 2008 Sep;180(1):301-16. doi: 10.1534/genetics.108.089706. Epub 2008 Aug 20. Genetics. 2008. PMID: 18716333 Free PMC article.
References
-
- Applied Biosystems. ABI PRISM GENESCAN Analysis Software 3.7. Applied Biosystems; Foster City, California: 2000.
-
- Barton NH. Clines in polygenic traits. Genetical Research. 1999;74:223–236. - PubMed
-
- Beaumont MA, Balding DJ. Identifying adaptive genetic divergence among populations from genome scans. Molecular Ecology. 2004;13:969–980. - PubMed
-
- Beaumont MA, Nichols RA. Evaluating loci for use in the genetic analysis of population structure. Proceedings of the Royal Society of London. Series B, Biological Sciences. 1996;263:1619–1626.
-
- Bradshaw HD, Schemske DW. Allele substitution at a flower colour locus produces a pollinator shift in monkeyflowers. Nature. 2003;426:176–178. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

