Serendipitous identification of natural intergenotypic recombinants of hepatitis C in Ireland
- PMID: 17107614
- PMCID: PMC1654145
- DOI: 10.1186/1743-422X-3-95
Serendipitous identification of natural intergenotypic recombinants of hepatitis C in Ireland
Abstract
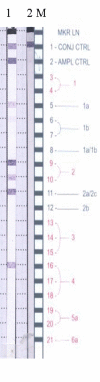
Background: Recombination between hepatitis C single stranded RNA viruses is a rare event. Natural viable intragenotypic and intergenotypic recombinants between 1b-1a, 1a-1c and 2k-1b, 2i-6p, respectively, have been reported. Diagnostically recombinants represent an intriguing challenge. Hepatitis C genotype is defined by interrogation of the sequence composition of the 5' untranslated region [5'UTR]. Occasionally, ambiguous specimens require further investigation of the genome, usually by interrogation of the NS5B region. The original purpose of this study was to confirm the existence of a suspected mixed genotype infection of genotypes 2 and 4 by clonal analysis at the NS5B region of the genome in two specimens from two separate individuals. This initial identification of genotype was based on analysis of the 5'UTR of the genome by reverse line probe hybridisation [RLPH].
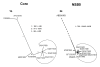
Results: The original diagnosis of a mixed genotype infection was not confirmed by clonal analysis of the NS5B region of the genome. The phylogenetic analysis indicated that both specimens were natural intergenotypic recombinant forms of HCV. The recombination was between genotypes 2k and 1b for both specimens. The recombination break point was identified as occurring within the NS2 region of the genome.
Conclusion: The viral recombinants identified here resemble the recombinant form originally identified in Russia. The RLPH pattern observed in this study may be a signature indicative of this particular type of intergenotype recombinant of hepatitis C meriting clonal analysis of NS2.
Figures

Similar articles
-
A natural intergenotypic recombinant of hepatitis C virus identified in St. Petersburg.J Virol. 2002 Apr;76(8):4034-43. doi: 10.1128/jvi.76.8.4034-4043.2002. J Virol. 2002. PMID: 11907242 Free PMC article.
-
Identification of hepatitis C virus 2k/1b intergenotypic recombinants in Georgia.Liver Int. 2018 Mar;38(3):451-457. doi: 10.1111/liv.13540. Epub 2017 Sep 2. Liver Int. 2018. PMID: 28782185
-
Natural intragenotypic and intergenotypic HCV recombinants are rare in southwest China even among patients with multiple exposures.J Clin Virol. 2010 Dec;49(4):272-6. doi: 10.1016/j.jcv.2010.08.007. Epub 2010 Sep 17. J Clin Virol. 2010. PMID: 20850377
-
Phylogenetic analysis of a circulating hepatitis C virus recombinant strain 1b/1a isolated in a French hospital centre.Infect Genet Evol. 2016 Jun;40:374-380. doi: 10.1016/j.meegid.2015.09.030. Epub 2015 Oct 9. Infect Genet Evol. 2016. PMID: 26444584
-
Novel nucleotide and amino acid covariation between the 5'UTR and the NS2/NS3 proteins of hepatitis C virus: bioinformatic and functional analyses.PLoS One. 2011;6(9):e25530. doi: 10.1371/journal.pone.0025530. Epub 2011 Sep 28. PLoS One. 2011. PMID: 21980483 Free PMC article.
Cited by
-
Recombination in hepatitis C virus.Viruses. 2011 Oct;3(10):2006-24. doi: 10.3390/v3102006. Epub 2011 Oct 24. Viruses. 2011. PMID: 22069526 Free PMC article. Review.
-
Mixed Infections Unravel Novel HCV Inter-Genotypic Recombinant Forms within the Conserved IRES Region.Viruses. 2024 Apr 3;16(4):560. doi: 10.3390/v16040560. Viruses. 2024. PMID: 38675902 Free PMC article.
-
New natural intergenotypic (2/5) recombinant of hepatitis C virus.J Virol. 2007 Apr;81(8):4357-62. doi: 10.1128/JVI.02639-06. Epub 2007 Jan 31. J Virol. 2007. PMID: 17267503 Free PMC article.
-
Origin and evolution of the unique hepatitis C virus circulating recombinant form 2k/1b.J Virol. 2012 Feb;86(4):2212-20. doi: 10.1128/JVI.06184-11. Epub 2011 Nov 23. J Virol. 2012. PMID: 22114341 Free PMC article.
-
Heads or Tails: Genotyping of Hepatitis C Virus Concerning the 2k/1b Circulating Recombinant Form.Int J Mol Sci. 2016 Aug 23;17(9):1384. doi: 10.3390/ijms17091384. Int J Mol Sci. 2016. PMID: 27563879 Free PMC article.
References
-
- Simmonds P, Holmes EC, Cha TA, Chan SW, McOmish F, Irvine B, Beall E, Yap PL, Kolberg J, Urdea MS. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J Gen Virol. 1993;74:2391–2399. - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

