Severe acute respiratory syndrome coronavirus protein 6 accelerates murine coronavirus infections
- PMID: 17108045
- PMCID: PMC1797517
- DOI: 10.1128/JVI.01515-06
Severe acute respiratory syndrome coronavirus protein 6 accelerates murine coronavirus infections
Abstract
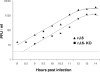
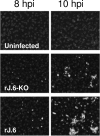
One or more of the unique 3'-proximal open reading frames (ORFs) of the severe acute respiratory syndrome (SARS) coronavirus may encode determinants of virus virulence. A prime candidate is ORF6, which encodes a 63-amino-acid membrane-associated peptide that can dramatically increase the lethality of an otherwise attenuated JHM strain of murine coronavirus (L. Pewe, H. Zhou, J. Netland, C. Tangudu, H. Olivares, L. Shi, D. Look, T. Gallagher, and S. Perlman, J. Virol. 79:11335-11342, 2005). To discern virulence mechanisms, we compared the in vitro growth properties of rJ.6, a recombinant JHM expressing the SARS peptide, with isogenic rJ.6-KO, which has an inactive ORF containing a mutated initiation codon and a termination codon at internal position 27. The rJ.6 infections proceeded rapidly, secreting progeny about 1.5 h earlier than rJ.6-KO infections did. The rJ.6 infections were also set apart by early viral protein accumulation and by robust expansion via syncytia, a characteristic feature of JHM virus dissemination. We found no evidence for protein 6 operating at the virus entry or assembly stage, as virions from either infection were indistinguishable. Rather, protein 6 appeared to operate by fostering viral RNA and protein synthesis, as RNA quantifications by reverse transcription-quantitative PCR revealed viral RNA levels in the rJ.6 cultures that were five to eight times higher than those lacking protein 6. Furthermore, protein 6 coimmunoprecipitated with viral RNAs and colocalized on cytoplasmic vesicles with replicating viral RNAs. The SARS coronavirus encodes a novel membrane protein 6 that can accelerate replication of a related mouse virus, a property that may explain its ability to increase in vivo virus virulence.
Figures


References
-
- Chan, W. S., C. Wu, S. C. Chow, T. Cheung, K. F. To, W. K. Leung, P. K. Chan, K. C. Lee, H. K. Ng, D. M. Au, and A. W. Lo. 2005. Coronaviral hypothetical and structural proteins were found in the intestinal surface enterocytes and pneumocytes of severe acute respiratory syndrome (SARS). Mod. Pathol. 18:1432-1439. - PMC - PubMed
-
- Choi, K. S., P. Huang, and M. M. Lai. 2002. Polypyrimidine-tract-binding protein affects transcription but not translation of mouse hepatitis virus RNA. Virology 303:58-68. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

