Mitochondria-associated yeast mRNAs and the biogenesis of molecular complexes
- PMID: 17108321
- PMCID: PMC1783778
- DOI: 10.1091/mbc.e06-09-0827
Mitochondria-associated yeast mRNAs and the biogenesis of molecular complexes
Abstract
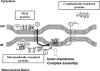
The coherence of mitochondrial biogenesis relies on spatiotemporally coordinated associations of 800-1000 proteins mostly encoded in the nuclear genome. We report the development of new quantitative analyses to assess the role of local protein translation in the construction of molecular complexes. We used real-time PCR to determine the cellular location of 112 mRNAs involved in seven mitochondrial complexes. Five typical cases were examined by an improved FISH protocol. The proteins produced in the vicinity of mitochondria (MLR proteins) were, almost exclusively, of prokaryotic origin and are key elements of the core construction of the molecular complexes; the accessory proteins were translated on free cytoplasmic polysomes. These two classes of proteins correspond, at least as far as intermembrane space (IMS) proteins are concerned, to two different import pathways. Import of MLR proteins involves both TOM and TIM23 complexes whereas non-MLR proteins only interact with the TOM complex. Site-specific translation loci, both outside and inside mitochondria, may coordinate the construction of molecular complexes composed of both nuclearly and mitochondrially encoded subunits.
Figures



Similar articles
-
The membrane insertase Oxa1 is required for efficient import of carrier proteins into mitochondria.J Mol Biol. 2012 Nov 2;423(4):590-9. doi: 10.1016/j.jmb.2012.07.018. Epub 2012 Jul 27. J Mol Biol. 2012. PMID: 22846909
-
Long mRNAs coding for yeast mitochondrial proteins of prokaryotic origin preferentially localize to the vicinity of mitochondria.Genome Biol. 2003;4(7):R44. doi: 10.1186/gb-2003-4-7-r44. Epub 2003 Jun 6. Genome Biol. 2003. PMID: 12844360 Free PMC article.
-
The outer membrane form of the mitochondrial protein Mcr1 follows a TOM-independent membrane insertion pathway.FEBS Lett. 2008 Mar 19;582(6):855-60. doi: 10.1016/j.febslet.2008.02.009. Epub 2008 Feb 13. FEBS Lett. 2008. PMID: 18279676
-
The enigmatic role of Mim1 in mitochondrial biogenesis.Eur J Cell Biol. 2010 Feb-Mar;89(2-3):212-5. doi: 10.1016/j.ejcb.2009.11.002. Epub 2009 Nov 26. Eur J Cell Biol. 2010. PMID: 19944477 Review.
-
Coupling of import and assembly pathways in mitochondrial protein biogenesis.Biol Chem. 2019 Dec 18;401(1):117-129. doi: 10.1515/hsz-2019-0310. Biol Chem. 2019. PMID: 31513529 Review.
Cited by
-
Nuclear expression of mitochondrial ND4 leads to the protein assembling in complex I and prevents optic atrophy and visual loss.Mol Ther Methods Clin Dev. 2015 Feb 25;2:15003. doi: 10.1038/mtm.2015.3. eCollection 2015. Mol Ther Methods Clin Dev. 2015. PMID: 26029714 Free PMC article.
-
Characterization of Factors Involved in Localized Translation Near Mitochondria by Ribosome-Proximity Labeling.Front Cell Dev Biol. 2019 Nov 26;7:305. doi: 10.3389/fcell.2019.00305. eCollection 2019. Front Cell Dev Biol. 2019. PMID: 31929983 Free PMC article.
-
CeFra-seq reveals broad asymmetric mRNA and noncoding RNA distribution profiles in Drosophila and human cells.RNA. 2018 Jan;24(1):98-113. doi: 10.1261/rna.063172.117. Epub 2017 Oct 27. RNA. 2018. PMID: 29079635 Free PMC article.
-
Metal acquisition and availability in the mitochondria.Chem Rev. 2009 Oct;109(10):4708-21. doi: 10.1021/cr900006y. Chem Rev. 2009. PMID: 19522505 Free PMC article. Review. No abstract available.
-
A genomic integration method for the simultaneous visualization of endogenous mRNAs and their translation products in living yeast.RNA. 2011 Dec;17(12):2249-55. doi: 10.1261/rna.029637.111. Epub 2011 Oct 24. RNA. 2011. PMID: 22025736 Free PMC article.
References
-
- Ackerman S. H., Tzagoloff A. Function, structure, and biogenesis of mitochondrial ATP synthase. Prog. Nucleic Acid Res. Mol. Biol. 2005;80:95–133. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

