Pol I transcription and pre-rRNA processing are coordinated in a transcription-dependent manner in mammalian cells
- PMID: 17108330
- PMCID: PMC1783775
- DOI: 10.1091/mbc.e06-03-0249
Pol I transcription and pre-rRNA processing are coordinated in a transcription-dependent manner in mammalian cells
Abstract
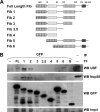
Pre-rRNA synthesis and processing are key steps in ribosome biogenesis. Although recent evidence in yeast suggests that these two processes are coupled, the nature of their association is unclear. In this report, we analyze the coordination between rDNA transcription and pre-rRNA processing in mammalian cells. We found that pol I transcription factor UBF interacts with pre-rRNA processing factors as analyzed by immunoprecipitations, and the association depends on active rRNA synthesis. In addition, injections of plasmids containing the human rDNA promoter and varying lengths of 18S rDNA into HeLa nuclei show that pol I transcription machinery can be recruited to rDNA promoters regardless of the product that is transcribed, whereas subgroups of pre-rRNA processing factors are recruited to plasmids only when specific pre-rRNA fragments are produced. Our observations suggest a model for sequential recruitment of pol I transcription factors and pre-rRNA processing factors to elongating pre-rRNA on an as-needed basis rather than corecruitment to sites of active transcription.
Figures
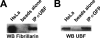
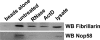


References
-
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K., editors. Short Protocols in Molecular Biology. New York: John Wiley & Sons; 2002.
-
- Bell S. P., Learned R. M., Jantzen H. M., Tjian R. Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science. 1988;241:1192–1197. - PubMed
-
- Bentley D. L. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr. Opin. Cell Biol. 2005;17:251–256. - PubMed
-
- Buratowski S. The CTD code. Nat. Struct. Biol. 2003;10:679–680. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

