Visual setup of logical models of signaling and regulatory networks with ProMoT
- PMID: 17109765
- PMCID: PMC1665465
- DOI: 10.1186/1471-2105-7-506
Visual setup of logical models of signaling and regulatory networks with ProMoT
Abstract
Background: The analysis of biochemical networks using a logical (Boolean) description is an important approach in Systems Biology. Recently, new methods have been proposed to analyze large signaling and regulatory networks using this formalism. Even though there is a large number of tools to set up models describing biological networks using a biochemical (kinetic) formalism, however, they do not support logical models.
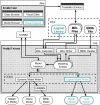
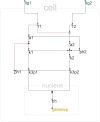
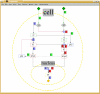
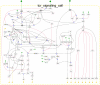
Results: Herein we present a flexible framework for setting up large logical models in a visual manner with the software tool ProMoT. An easily extendible library, ProMoT's inherent modularity and object-oriented concept as well as adaptive visualization techniques provide a versatile environment. Both the graphical and the textual description of the logical model can be exported to different formats.
Conclusion: New features of ProMoT facilitate an efficient set-up of large Boolean models of biochemical interaction networks. The modeling environment is flexible; it can easily be adapted to specific requirements, and new extensions can be introduced. ProMoT is freely available from http://www.mpi-magdeburg.mpg.de/projects/promot/.
Figures


References
-
- Thomas R, D'Ari R. Biological Feedback. Boca Raton, CRC Press; 1990.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

