Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism
- PMID: 17110578
- PMCID: PMC2754788
- DOI: 10.1126/science.1131399
Initial transcription by RNA polymerase proceeds through a DNA-scrunching mechanism
Abstract
Using fluorescence resonance energy transfer to monitor distances within single molecules of abortively initiating transcription initiation complexes, we show that initial transcription proceeds through a "scrunching" mechanism, in which RNA polymerase (RNAP) remains fixed on promoter DNA and pulls downstream DNA into itself and past its active center. We show further that putative alternative mechanisms for RNAP active-center translocation in initial transcription, involving "transient excursions" of RNAP relative to DNA or "inchworming" of RNAP relative to DNA, do not occur. The results support a model in which a stressed intermediate, with DNA-unwinding stress and DNA-compaction stress, is formed during initial transcription, and in which accumulated stress is used to drive breakage of interactions between RNAP and promoter DNA and between RNAP and initiation factors during promoter escape.
Figures

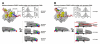

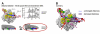
Comment in
-
Biochemistry. RNA polymerase, a scrunching machine.Science. 2006 Nov 17;314(5802):1097-8. doi: 10.1126/science.1135746. Science. 2006. PMID: 17110563 No abstract available.
References
-
- Record MTJ, Reznikoff W, Craig M, McQuade K, Schlax P. In: Escherichia coli and Salmonella. Neidhart F, editor. vol. 1. ASM Press; Washington, D.C.: 1996. pp. 792–820.
-
- Young B, Gruber T, Gross C. Cell. 2002;109:417–420. - PubMed
-
- Murakami K, Darst S. Curr. Opin. Struct. Biol. 2003;13:31–39. - PubMed
-
- Hsu L. Biochim. Biophys. Acta. 2002;1577:191–207. - PubMed
-
- Carpousis A, Gralla J. J. Mol. Biol. 1985;183:165–177. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

