Indoleamine 2,3-dioxygenase-expressing dendritic cells form suppurative granulomas following Listeria monocytogenes infection
- PMID: 17111046
- PMCID: PMC1636691
- DOI: 10.1172/JCI28996
Indoleamine 2,3-dioxygenase-expressing dendritic cells form suppurative granulomas following Listeria monocytogenes infection
Abstract
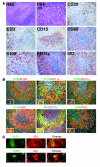
Control of pathogens by formation of abscesses and granulomas is a major strategy of the innate immune system, especially when effector mechanisms of adaptive immunity are insufficient. We show in human listeriosis that DCs expressing indoleamine 2,3-dioxygenase (IDO), together with macrophages, are major cellular components of suppurative granulomas in vivo. Induction of IDO by DCs is a cell-autonomous response to Listeria monocytogenes infection and was also observed in other granulomatous infections with intracellular bacteria, such as Bartonella henselae. Reporting on our use of the clinically applied anti-TNF-alpha antibody infliximab, we further demonstrate in vitro that IDO induction is TNF-alpha dependent. Repression of IDO therefore might result in exacerbation of granulomatous diseases observed during anti-TNF-alpha therapy. These findings place IDO(+) DCs not only at the intersection of innate and adaptive immunity but also at the forefront of bacterial containment in granulomatous infections.
Figures

References
-
- Kaufmann S.H. Immunity to intracellular bacteria. Annu. Rev. Immunol. 1993;11:129–163. - PubMed
-
- Gordon S. Alternative activation of macrophages. Nat. Rev. Immunol. 2003;3:23–35. - PubMed
-
- Pamer E.G. Immune responses to Listeria monocytogenes. Nat. Rev. Immunol. 2004;4:812–823. - PubMed
-
- Unanue E.R. Studies in listeriosis show the strong symbiosis between the innate cellular system and the T-cell response. Immunol. Rev. 1997;158:11–25. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

