Epitope mapping and topographic analysis of VAR2CSA DBL3X involved in P. falciparum placental sequestration
- PMID: 17112315
- PMCID: PMC1636682
- DOI: 10.1371/journal.ppat.0020124
Epitope mapping and topographic analysis of VAR2CSA DBL3X involved in P. falciparum placental sequestration
Abstract
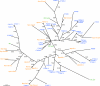
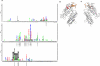
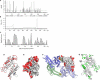
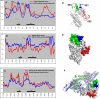
Pregnancy-associated malaria is a major health problem, which mainly affects primigravidae living in malaria endemic areas. The syndrome is precipitated by accumulation of infected erythrocytes in placental tissue through an interaction between chondroitin sulphate A on syncytiotrophoblasts and a parasite-encoded protein on the surface of infected erythrocytes, believed to be VAR2CSA. VAR2CSA is a polymorphic protein of approximately 3,000 amino acids forming six Duffy-binding-like (DBL) domains. For vaccine development it is important to define the antigenic targets for protective antibodies and to characterize the consequences of sequence variation. In this study, we used a combination of in silico tools, peptide arrays, and structural modeling to show that sequence variation mainly occurs in regions under strong diversifying selection, predicted to form flexible loops. These regions are the main targets of naturally acquired immunoglobulin gamma and accessible for antibodies reacting with native VAR2CSA on infected erythrocytes. Interestingly, surface reactive anti-VAR2CSA antibodies also target a conserved DBL3X region predicted to form an alpha-helix. Finally, we could identify DBL3X sequence motifs that were more likely to occur in parasites isolated from primi- and multigravidae, respectively. These findings strengthen the vaccine candidacy of VAR2CSA and will be important for choosing epitopes and variants of DBL3X to be included in a vaccine protecting women against pregnancy-associated malaria.
Conflict of interest statement
Figures



References
-
- Christophers SR. The mechanism of immunity against malaria in communities living under hyper-endemic conditions. Ind J Med Res. 1924;12:273–294.
-
- Bray RS, Anderson MJ. Falciparum malaria and pregnancy. Trans R Soc Trop Med Hyg. 1979;73:427–431. - PubMed
-
- McGregor IA, Wilson ME, Billewicz WZ. Malaria infection of the placenta in Gambia, West Africa: Its incidence and relationship to stillbirth, birth-weight, and placental weight. Trans R Soc Trop Med Hyg. 1983;77:232–244. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

