Integration and mining of malaria molecular, functional and pharmacological data: how far are we from a chemogenomic knowledge space?
- PMID: 17112376
- PMCID: PMC1665468
- DOI: 10.1186/1475-2875-5-110
Integration and mining of malaria molecular, functional and pharmacological data: how far are we from a chemogenomic knowledge space?
Abstract
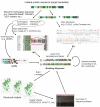
The organization and mining of malaria genomic and post-genomic data is important to significantly increase the knowledge of the biology of its causative agents, and is motivated, on a longer term, by the necessity to predict and characterize new biological targets and new drugs. Biological targets are sought in a biological space designed from the genomic data from Plasmodium falciparum, but using also the millions of genomic data from other species. Drug candidates are sought in a chemical space containing the millions of small molecules stored in public and private chemolibraries. Data management should, therefore, be as reliable and versatile as possible. In this context, five aspects of the organization and mining of malaria genomic and post-genomic data were examined: 1) the comparison of protein sequences including compositionally atypical malaria sequences, 2) the high throughput reconstruction of molecular phylogenies, 3) the representation of biological processes, particularly metabolic pathways, 4) the versatile methods to integrate genomic data, biological representations and functional profiling obtained from X-omic experiments after drug treatments and 5) the determination and prediction of protein structures and their molecular docking with drug candidate structures. Recent progress towards a grid-enabled chemogenomic knowledge space is discussed.
Figures



References
-
- World Malaria Report 2005. Geneva, World Health Organization, WHO/UNICEF. 2005.
-
- Adl SM, Simpson AG, Farmer MA, Andersen RA, Anderson OR, Barta JR, Bowser SS, Brugerolle G, Fensome RA, Fredericq S, James TY, Karpov S, Kugrens P, Krug J, Lane CE, Lewis LA, Lodge J, Lynn DH, Mann DG, McCourt RM, Mendoza L, Moestrup O, Mozley-Standridge SE, Nerad TA, Shearer CA, Smirnov AV, Spiegel FW, Taylor MF. The new higher level classification of eukaryotes with emphasis on the taxonomy of protists. J Eukaryot Microbiol. 2005;52:399–351. - PubMed
-
- Desowitz RS. Malaria: from quinine to the vaccine. Hosp Pract. 1992;27:209–14. 217–24, 229–32. - PubMed
-
- Utzinger J, Tanner M, Kammen DM, Killeen GF, Singer BH. Integrated program is key to malarial control. Nature. 2002;419:431. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

