Characterizing 56 complete SARS-CoV S-gene sequences from Hong Kong
- PMID: 17112780
- PMCID: PMC7108452
- DOI: 10.1016/j.jcv.2006.10.001
Characterizing 56 complete SARS-CoV S-gene sequences from Hong Kong
Abstract
Background: The spike glycoprotein (S) gene of the severe acute respiratory syndrome-associated coronavirus (SARS-CoV) has been useful in analyzing the molecular epidemiology of the 2003 SARS outbreaks.
Objectives: To characterize complete SARS-CoV S-gene sequences from Hong Kong.
Study design: Fifty-six SARS-CoV S-gene sequences, obtained from patients who presented with SARS to the Prince of Wales Hospital during March-May 2003, were analysed using a maximum likelihood (ML) approach, together with 138 other (both human and animal) S-gene sequences downloaded from GenBank.
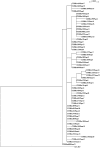
Results: The maximum-likelihood (ML) trees showed little evolution occurring within these 56 sequences. Analysis with the other sequences, showed three distinct SARS clusters, closely correlated to previously defined early, middle and late phases of the 2003 international SARS outbreaks. In addition, two new single nucleotide variations (SNVs), T21615A and T21901A, were discovered, not previously reported elsewhere.
Conclusions: The ML approach to the reconstruction of tree phylogenies is known to be superior to the more popular, less computationally and time-demanding neighbour-joining (NJ) approach. The ML analysis in this study confirms the previously reported SARS epidemiology analysed mostly using the NJ approach. The two new SNVs reported here are most likely due to the tissue-culture passaging of the clinical samples.
Figures


References
-
- Chinese, SARS Molecular Epidemiology Consortium Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science. 2004;303(5664):1666–1669. - PubMed
-
- Guan Y., Zheng B.J., He Y.Q. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302(5643):276–278. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

