FRET study of membrane proteins: determination of the tilt and orientation of the N-terminal domain of M13 major coat protein
- PMID: 17114224
- PMCID: PMC1783881
- DOI: 10.1529/biophysj.106.095026
FRET study of membrane proteins: determination of the tilt and orientation of the N-terminal domain of M13 major coat protein
Abstract
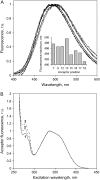
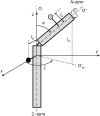
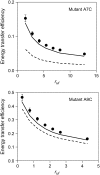
A formalism for membrane protein structure determination was developed. This method is based on steady-state FRET data and information about the position of the fluorescence maxima on site-directed fluorescent labeled proteins in combination with global data analysis utilizing simulation-based fitting. The methodology was applied to determine the structural properties of the N-terminal domain of the major coat protein from bacteriophage M13 reconstituted into unilamellar DOPC/DOPG (4:1 mol/mol) vesicles. For our purpose, the cysteine mutants A7C, A9C, N12C, S13C, Q15C, A16C, S17C, and A18C in the N-terminal domain of this protein were produced and specifically labeled with the fluorescence probe AEDANS. The energy transfer data from the natural Trp-26 to AEDANS were analyzed assuming a two-helix protein model. Furthermore, the polarity Stokes shift of the AEDANS fluorescence maxima is taken into account. As a result the orientation and tilt of the N-terminal protein domain with respect to the bilayer interface were obtained, showing for the first time, to our knowledge, an overall alpha-helical protein conformation from amino acid residues 12-46, close to the protein conformation in the intact phage.
Figures


Similar articles
-
FRET study of membrane proteins: simulation-based fitting for analysis of membrane protein embedment and association.Biophys J. 2006 Jul 15;91(2):454-66. doi: 10.1529/biophysj.106.082867. Epub 2006 Apr 21. Biophys J. 2006. PMID: 16632512 Free PMC article.
-
Lipid bilayer topology of the transmembrane alpha-helix of M13 Major coat protein and bilayer polarity profile by site-directed fluorescence spectroscopy.Biophys J. 2004 Sep;87(3):1445-55. doi: 10.1529/biophysj.104.043208. Biophys J. 2004. PMID: 15345527 Free PMC article.
-
Membrane assembly of M13 major coat protein: evidence for a structural adaptation in the hinge region and a tilted transmembrane domain.Biochemistry. 2004 Nov 9;43(44):13972-80. doi: 10.1021/bi048437x. Biochemistry. 2004. PMID: 15518546
-
From 'I' to 'L' and back again: the odyssey of membrane-bound M13 protein.Trends Biochem Sci. 2009 May;34(5):249-55. doi: 10.1016/j.tibs.2009.01.007. Epub 2009 Apr 8. Trends Biochem Sci. 2009. PMID: 19362002 Review.
-
Protein-lipid interactions of bacteriophage M13 gene 9 minor coat protein.Mol Membr Biol. 2004 Nov-Dec;21(6):351-9. doi: 10.1080/09687860400012918. Mol Membr Biol. 2004. PMID: 15764365 Review.
Cited by
-
Fluorescent Probes and Quenchers in Studies of Protein Folding and Protein-Lipid Interactions.Chem Rec. 2024 Feb;24(2):e202300232. doi: 10.1002/tcr.202300232. Epub 2023 Sep 11. Chem Rec. 2024. PMID: 37695081 Free PMC article. Review.
-
Viruses: incredible nanomachines. New advances with filamentous phages.Eur Biophys J. 2010 Mar;39(4):541-50. doi: 10.1007/s00249-009-0523-0. Epub 2009 Aug 13. Eur Biophys J. 2010. PMID: 19680644 Free PMC article. Review.
-
Orientation and dynamics of transmembrane peptides: the power of simple models.Eur Biophys J. 2010 Mar;39(4):609-21. doi: 10.1007/s00249-009-0567-1. Epub 2009 Dec 18. Eur Biophys J. 2010. PMID: 20020122 Free PMC article. Review.
-
Structure of membrane-embedded M13 major coat protein is insensitive to hydrophobic stress.Biophys J. 2007 Nov 15;93(10):3541-7. doi: 10.1529/biophysj.107.112698. Epub 2007 Aug 17. Biophys J. 2007. PMID: 17704180 Free PMC article.
-
Visualization of Protein Interactions in Living Cells.Self Nonself. 2011 Apr;2(2):98-107. doi: 10.4161/self.2.2.17932. Epub 2011 Apr 1. Self Nonself. 2011. PMID: 22299061 Free PMC article.
References
-
- Stopar, D., R. B. Spruijt, and M. A. Hemminga. 2006. Anchoring mechanisms of membrane-associated M13 major coat protein. Chem. Phys. Lipids. 141:83–93. - PubMed
-
- Vos, W. L., R. B. M. Koehorst, R. B. Spruijt, and M. A. Hemminga. 2005. Membrane-bound conformation of M13 major coat protein: a structure validations through FRET-derived constrains. J. Biol. Chem. 280:38522–38527. - PubMed
-
- Spruijt, R. B., A. B. Meijer, C. J. A. M. Wolfs, and M. A. Hemminga. 2000. Localization and rearrangement modulation of the N-terminal arm of the membrane-bound major coat protein of bacteriophage M13. Biochim. Biophys. Acta. 1509:311–323. - PubMed
-
- Spruijt, R. B., C. J. A. M. Wolfs, and M. A. Hemminga. 1989. Aggregation-related conformational change of the membrane-associated coat protein of bacteriophage M13. Biochemistry. 28:9158–9165. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

