Physical and functional interactions between Escherichia coli MutY glycosylase and mismatch repair protein MutS
- PMID: 17114250
- PMCID: PMC1797285
- DOI: 10.1128/JB.01513-06
Physical and functional interactions between Escherichia coli MutY glycosylase and mismatch repair protein MutS
Abstract
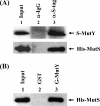
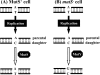
Escherichia coli MutY and MutS increase replication fidelity by removing adenines that were misincorporated opposite 7,8-dihydro-8-oxo-deoxyguanines (8-oxoG), G, or C. MutY DNA glycosylase removes adenines from these mismatches through a short-patch base excision repair pathway and thus prevents G:C-to-T:A and A:T-to-G:C mutations. MutS binds to the mismatches and initiates the long-patch mismatch repair on daughter DNA strands. We have previously reported that the human MutY homolog (hMYH) physically and functionally interacts with the human MutS homolog, hMutSalpha (Y. Gu et al., J. Biol. Chem. 277:11135-11142, 2002). Here, we show that a similar relationship between MutY and MutS exists in E. coli. The interaction of MutY and MutS involves the Fe-S domain of MutY and the ATPase domain of MutS. MutS, in eightfold molar excess over MutY, can enhance the binding activity of MutY with an A/8-oxoG mismatch by eightfold. The MutY expression level and activity in mutS mutant strains are sixfold and twofold greater, respectively, than those for the wild-type cells. The frequency of A:T-to-G:C mutations is reduced by two- to threefold in a mutS mutY mutant compared to a mutS mutant. Our results suggest that MutY base excision repair and mismatch repair defend against the mutagenic effect of 8-oxoG lesions in a cooperative manner.
Figures



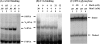

References
-
- Al-Tassan, N., N. H. Chmiel, J. Maynard, N. Fleming, A. L. Livingston, G. T. Williams, A. K. Hodges, D. R. Davies, S. S. David, J. R. Sampson, and J. P. Cheadle. 2002. Inherited variants of MYH associated with somatic G:C to T:A mutations in colorectal tumors. Nat. Genet. 30:227-232. - PubMed
-
- Biswas, I., G. Obmolova, M. Takahashi, A. Herr, M. A. Newman, W. Yang, and P. Hsieh. 2001. Disruption of the helix-U-turn-helix motif of MutS protein: loss of subunit dimerization, mismatch binding and ATP hydrolysis. J. Mol. Biol. 305:805-816. - PubMed
-
- Bjornson, K. P., L. J. Blackwell, H. Sage, C. Baitinger, D. Allen, and P. Modrich. 2003. Assembly and molecular activities of the MutS tetramer. J. Biol. Chem. 278:34667-34673. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

