Identification of diverse archaeal proteins with class III signal peptides cleaved by distinct archaeal prepilin peptidases
- PMID: 17114255
- PMCID: PMC1797317
- DOI: 10.1128/JB.01547-06
Identification of diverse archaeal proteins with class III signal peptides cleaved by distinct archaeal prepilin peptidases
Abstract
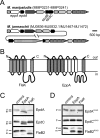
Most secreted archaeal proteins are targeted to the membrane via a tripartite signal composed of a charged N terminus and a hydrophobic domain, followed by a signal peptidase-processing site. Signal peptides of archaeal flagellins, similar to class III signal peptides of bacterial type IV pilins, are distinct in that their processing sites precede the hydrophobic domain, which is crucial for assembly of these extracytoplasmic structures. To identify the complement of archaeal proteins with class III signal sequences, a PERL program (FlaFind) was written. A diverse set of proteins was identified, and many of these FlaFind positives were encoded by genes that were cotranscribed with homologs of pilus assembly genes. Moreover, structural conservation of primary sequences between many FlaFind positives and subunits of bacterial pilus-like structures, which have been shown to be critical for pilin assembly, have been observed. A subset of pilin-like FlaFind positives contained a conserved domain of unknown function (DUF361) within the signal peptide. Many of the genes encoding these proteins were in operons that contained a gene encoding a novel euryarchaeal prepilin-peptidase, EppA, homolog. Heterologous analysis revealed that Methanococcus maripaludis DUF361-containing proteins were specifically processed by the EppA homolog of this archaeon. Conversely, M. maripaludis preflagellins were cleaved only by the archaeal preflagellin peptidase FlaK. Together, the results reveal a diverse set of archaeal proteins with class III signal peptides that might be subunits of as-yet-undescribed cell surface structures, such as archaeal pili.
Figures


References
-
- Albers, S. V., and A. J. Driessen. 2005. Analysis of ATPases of putative secretion operons in the thermoacidophilic archaeon Sulfolobus solfataricus. Microbiology 151:763-773. - PubMed
-
- Albers, S. V., and A. J. M. Driessen. 2002. Signal peptides of secreted proteins of the archaeon Sulfolobus solfataricus: a genomic survey. Arch. Microbiol. 177:209-216. - PubMed
-
- Albers, S. V., Z. Szabo, and A. J. Driessen. 2006. Protein secretion in the Archaea: multiple paths towards a unique cell surface. Nat. Rev. Microbiol. 4:537-547. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

