AtERF14, a member of the ERF family of transcription factors, plays a nonredundant role in plant defense
- PMID: 17114278
- PMCID: PMC1761963
- DOI: 10.1104/pp.106.086637
AtERF14, a member of the ERF family of transcription factors, plays a nonredundant role in plant defense
Abstract
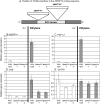
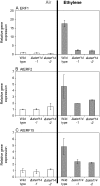
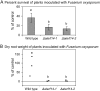
We had previously shown that several transcription factors of the ethylene (ET) response factor (ERF) family were induced with different but overlapping kinetics following challenge of Arabidopsis (Arabidopsis thaliana) with Pseudomonas syringae pv tomato DC3000 (avrRpt2). One of these genes, a transcriptional activator, AtERF14, was induced at the same time as ERF-target genes (ChiB, basic chitinase). To unravel the potential function of AtERF14 in regulating the plant defense response, we have analyzed gain- and loss-of-function mutants. We show here that AtERF14 has a prominent role in the plant defense response, since overexpression of AtERF14 had dramatic effects on both plant phenotype and defense gene expression and AtERF14 loss-of-function mutants showed impaired induction of defense genes following exogenous ET treatment and increased susceptibility to Fusarium oxysporum. Moreover, the expression of other ERF genes involved in defense and ET/jasmonic acid responses, such as ERF1 and AtERF2, depends on AtERF14 expression. A number of ERFs have been shown to function in the defense response through overexpression. However, the effect of loss of AtERF14 function on defense gene expression, pathogen resistance, and regulation of the expression of other ERF genes is unique thus far. These results suggest a unique role for AtERF14 in regulating the plant defense response.
Figures





References
-
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 - PubMed
-
- Anderson JP, Badruzsaufari E, Schenk PM, Manners JM, Desmond OJ, Ehlert C, Maclean DJ, Ebert PR, Kazan K (2004) Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16 3460–3479 - PMC - PubMed
-
- Berrocal-Lobo M, Molina A, Solano R (2002) Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J 29 23–32 - PubMed
-
- Booth C (1977) Fusarium: Laboratory Guide to the Identification of the Major Species. Kew Commonwealth Mycological Institute, Surrey, UK
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

