Psb27, a cyanobacterial lipoprotein, is involved in the repair cycle of photosystem II
- PMID: 17114356
- PMCID: PMC1693947
- DOI: 10.1105/tpc.106.042671
Psb27, a cyanobacterial lipoprotein, is involved in the repair cycle of photosystem II
Abstract
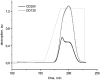
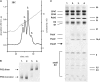
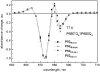
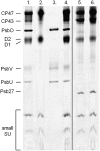
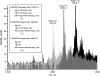
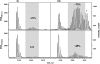
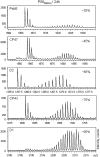
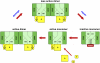
Photosystem II (PSII) performs one of the key reactions on our planet: the light-driven oxidation of water. This fundamental but very complex process requires PSII to act in a highly coordinated fashion. Despite detailed structural information on the fully assembled PSII complex, the dynamic aspects of formation, processing, turnover, and degradation of PSII with at least 19 subunits and various cofactors are still not fully understood. Transient complexes are especially difficult to characterize due to low abundance, potential heterogeneity, and instability. Here, we show that Psb27 is involved in the assembly of the water-splitting site of PSII and in the turnover of the complex. Psb27 is a bacterial lipoprotein with a specific lipid modification as shown by matrix-assisted laser-desorption ionization time of flight mass spectrometry. The combination of HPLC purification of four different PSII subcomplexes and (15)N pulse label experiments revealed that lipoprotein Psb27 is part of a preassembled PSII subcomplex that represents a distinct intermediate in the repair cycle of PSII.
Figures

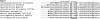
References
-
- Anderson, J.M., Park, Y.I., and Chow, W.S. (1997). Photoinactivation and photoprotection of photosystem II in nature. Physiol. Plant 100 214–223.
-
- Aro, E.M., Suorsa, M., Rokka, A., Allahverdiyeva, Y., Paakkarinen, V., Saleem, A., Battchikova, N., and Rintamaki, E. (2005). Dynamics of photosystem II: A proteomic approach to thylakoid protein complexes. J. Exp. Bot. 56 347–356. - PubMed
-
- Bendtsen, J.D., Nielsen, H., von Heijne, G., and Brunak, S. (2004). Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340 783–795. - PubMed
-
- Chen, H., Zhang, D., Guo, J., Wu, H., Jin, M., Lu, Q., Lu, C., and Zhang, L. (2006). A Psb27 homologue in Arabidopsis thaliana is required for efficient repair of photodamaged photosystem II. Plant Mol. Biol. 61 567–575. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

