MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula
- PMID: 17114582
- PMCID: PMC1635144
- DOI: 10.1101/gad.402806
MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula
Abstract
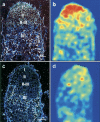
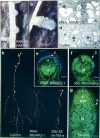
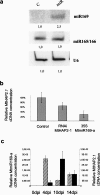
In the model legume Medicago truncatula, we identified a new transcription factor of the CCAAT-binding family, MtHAP2-1, for which RNA interference (RNAi) and in situ hybridization experiments indicate a key role during nodule development, possibly by controlling nodule meristem function. We could also show that MtHAP2-1 is regulated by microRNA169, whose overexpression leads to the same nodule developmental block as MtHAP2-1 RNAi constructs. The complementary expression pattern of miR169 and MtHAP2-1 and the phenotype of miR169-resistant MtHAP2-1 nodules strongly suggest, in addition, that the miR169-mediated restriction of MtHAP2-1 expression to the nodule meristematic zone is essential for the differentiation of nodule cells.
Figures


References
-
- Ardourel, M., Demont, N., Debelle, F., Maillet, F., de Billy, F., Prome, J.C., Denarie, J., Truchet, G. Rhizobium meliloti lipooligosaccharide nodulation factors: Different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell. 1994;6:1357–1374. - PMC - PubMed
-
- Ben-Naim, O., Eshed, R., Parnis, A., Teper-Bamnolker, P., Shalit, A., Coupland, G., Samach, A., Lifschitz, E. The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J. 2006;46:462–476. - PubMed
-
- Bhattacharya, A., Deng, J.M., Zhang, Z., Behringer, R., de Crombrugghe, B., Maity, S.N. The B subunit of the CCAAT box binding transcription factor complex (CBF/NF-Y) is essential for early mouse development and cell proliferation. Cancer Res. 2003;63:8167–8172. - PubMed
-
- Boisson-Dernier, A., Andriankaja, A., Chabaud, M., Niebel, A., Journet, E.P., Barker, D.G., de Carvalho-Niebel, F. MtENOD11 gene activation during rhizobial infection and mycorrhizal arbuscule development requires a common AT-rich-containing regulatory sequence. Mol. Plant Microbe Interact. 2005;18:1269–1276. - PubMed
-
- El Yahyaoui, F., Kuster, H., Ben Amor, B., Hohnjec, N., Pühler, A., Becker, A., Gouzy, J., Vernié, T., Gough, C., Niebel, A., et al. Expression profiling in Medicago truncatula identifies more than 750 genes differentially expressed during nodulation, including many potential regulators of the symbiotic program. Plant Physiol. 2004;136:3559–3576. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
