Mechanism of Dun1 activation by Rad53 phosphorylation in Saccharomyces cerevisiae
- PMID: 17114794
- PMCID: PMC2811688
- DOI: 10.1074/jbc.M609322200
Mechanism of Dun1 activation by Rad53 phosphorylation in Saccharomyces cerevisiae
Abstract
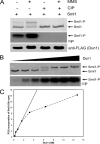
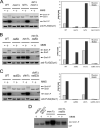
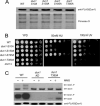
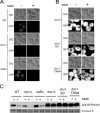
Despite extensive studies, the molecular mechanism of DNA damage checkpoint activation remains incompletely understood. To better dissect this mechanism, we developed an activity-based assay for Dun1, a downstream DNA damage check-point kinase in yeast, using its physiological substrate Sml1. Using this assay, we confirmed the genetic basis of Dun1 activation. Rad53 was found to be directly responsible for Dun1 activation. We reconstituted the activation of Dun1 by Rad53 and found that phosphorylation of Thr-380 in the activation loop of Dun1 by Rad53 is responsible for Dun1 activation. Interestingly, phosphorylation of the evolutionarily conserved Thr-354 in the activation loop of Rad53 is also important for the regulation of Rad53 activity. Thus, this conserved mode of activation loop phosphorylation appears to be a general mechanism for the activation of Chk2 family kinases.
Figures


References
-
- Nyberg KA, Michelson RJ, Putnam CW, Weinert TA. Annu. Rev. Genet. 2002;36:617–656. - PubMed
-
- Jackson SP. Curr. Opin. Genet. Dev. 1996;6:19–25. - PubMed
-
- Sanchez Y, Desany BA, Jones WJ, Liu Q, Wang B, Elledge SJ. Science. 1996;271:357–360. - PubMed
-
- Alcasabas AA, Osborn AJ, Bachant J, Hu F, Werler PJ, Bousset K, Furuya K, Diffley JF, Carr AM, Elledge SJ. Nat. Cell Biol. 2001;3:958–965. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

