Mathematical modeling of intracellular signaling pathways
- PMID: 17118154
- PMCID: PMC1775040
- DOI: 10.1186/1471-2202-7-S1-S10
Mathematical modeling of intracellular signaling pathways
Abstract
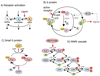
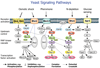
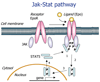
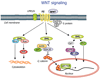
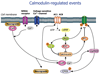
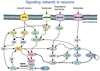
Dynamic modeling and simulation of signal transduction pathways is an important topic in systems biology and is obtaining growing attention from researchers with experimental or theoretical background. Here we review attempts to analyze and model specific signaling systems. We review the structure of recurrent building blocks of signaling pathways and their integration into more comprehensive models, which enables the understanding of complex cellular processes. The variety of mechanisms found and modeling techniques used are illustrated with models of different signaling pathways. Focusing on the close interplay between experimental investigation of pathways and the mathematical representations of cellular dynamics, we discuss challenges and perspectives that emerge in studies of signaling systems.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

