A posterior centre establishes and maintains polarity of the Caenorhabditis elegans embryo by a Wnt-dependent relay mechanism
- PMID: 17121454
- PMCID: PMC1637133
- DOI: 10.1371/journal.pbio.0040396
A posterior centre establishes and maintains polarity of the Caenorhabditis elegans embryo by a Wnt-dependent relay mechanism
Abstract
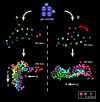
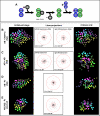
Cellular polarity is a general feature of animal development. However, the mechanisms that establish and maintain polarity in a field of cells or even in the whole embryo remain elusive. Here we provide evidence that in the Caenorhabditis elegans embryo, the descendants of P1, the posterior blastomere of the 2-cell stage, constitute a polarising centre that orients the cell divisions of most of the embryo. This polarisation depends on a MOM-2/Wnt signal originating from the P1 descendants. Furthermore, we show that the MOM-2/Wnt signal is transduced from cell to cell by a relay mechanism. Our findings suggest how polarity is first established and then maintained in a field of cells. According to this model, the relay mechanism constantly orients the polarity of all cells towards the polarising centre, thus organising the whole embryo. This model may also apply to other systems such as Drosophila and vertebrates.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures



Comment in
-
Back to Front in C. elegans.PLoS Biol. 2006 Dec;4(12):e426. doi: 10.1371/journal.pbio.0040426. Epub 2006 Nov 21. PLoS Biol. 2006. PMID: 20076511 Free PMC article. No abstract available.
References
-
- Keller R. Shaping the vertebrate body plan by polarized embryonic cell movements. Science. 2002;298:1950–1954. - PubMed
-
- Baena-Lopez LA, Baonza A, Garcia-Bellido A. The orientation of cell divisions determines the shape of Drosophila organs. Curr Biol. 2005;15:1640–1644. - PubMed
-
- Gong Y, Mo C, Fraser SE. Planar cell polarity signalling controls cell division orientation during zebrafish gastrulation. Nature. 2004;430:689–693. - PubMed
-
- Brownlee C, Bouget F-Y, Corellou F. Choosing sides: Establishment of polarity in zygotes of fucoid algae. Seminars Cell Dev Biol. 2001;12:343–351. - PubMed
-
- Labbe JC, Goldstein B. Embryonic development: A new SPN on cell fate specification. Curr Biol. 2002;12:R396–398. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

