Directed evolution and structural analysis of N-carbamoyl-D-amino acid amidohydrolase provide insights into recombinant protein solubility in Escherichia coli
- PMID: 17121498
- PMCID: PMC1863561
- DOI: 10.1042/BJ20061457
Directed evolution and structural analysis of N-carbamoyl-D-amino acid amidohydrolase provide insights into recombinant protein solubility in Escherichia coli
Abstract
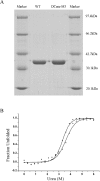
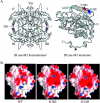
One of the greatest bottlenecks in producing recombinant proteins in Escherichia coli is that over-expressed target proteins are mostly present in an insoluble form without any biological activity. DCase (N-carbamoyl-D-amino acid amidohydrolase) is an important enzyme involved in semi-synthesis of beta-lactam antibiotics in industry. In the present study, in order to determine the amino acid sites responsible for solubility of DCase, error-prone PCR and DNA shuffling techniques were applied to randomly mutate its coding sequence, followed by an efficient screening based on structural complementation. Several mutants of DCase with reduced aggregation were isolated. Solubility tests of these and several other mutants generated by site-directed mutagenesis indicated that three amino acid residues of DCase (Ala18, Tyr30 and Lys34) are involved in its protein solubility. In silico structural modelling analyses suggest further that hydrophilicity and/or negative charge at these three residues may be responsible for the increased solubility of DCase proteins in E. coli. Based on this information, multiple engineering designated mutants were constructed by site-directed mutagenesis, among them a triple mutant A18T/Y30N/K34E (named DCase-M3) could be overexpressed in E. coli and up to 80% of it was soluble. DCase-M3 was purified to homogeneity and a comparative analysis with wild-type DCase demonstrated that DCase-M3 enzyme was similar to the native DCase in terms of its kinetic and thermodynamic properties. The present study provides new insights into recombinant protein solubility in E. coli.
Figures
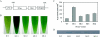


Similar articles
-
Gradually accumulating beneficial mutations to improve the thermostability of N-carbamoyl-D-amino acid amidohydrolase by step-wise evolution.Appl Microbiol Biotechnol. 2011 May;90(4):1361-71. doi: 10.1007/s00253-011-3114-9. Epub 2011 Mar 1. Appl Microbiol Biotechnol. 2011. PMID: 21360152
-
Improving the thermostability of N-carbamyl-D-amino acid amidohydrolase by error-prone PCR.Appl Microbiol Biotechnol. 2009 Feb;82(2):279-85. doi: 10.1007/s00253-008-1748-z. Epub 2008 Nov 5. Appl Microbiol Biotechnol. 2009. PMID: 18985337
-
Functional expression of a plant hydroxynitrile lyase in Escherichia coli by directed evolution: creation and characterization of highly in vivo soluble mutants.Protein Eng Des Sel. 2011 Aug;24(8):607-16. doi: 10.1093/protein/gzr030. Epub 2011 Jul 5. Protein Eng Des Sel. 2011. PMID: 21729945
-
Predicting the solubility of recombinant proteins in Escherichia coli.Methods Mol Biol. 2015;1258:403-8. doi: 10.1007/978-1-4939-2205-5_23. Methods Mol Biol. 2015. PMID: 25447878 Review.
-
Engineering of L-amino acid deaminases for the production of α-keto acids from L-amino acids.Bioengineered. 2019 Dec;10(1):43-51. doi: 10.1080/21655979.2019.1595990. Bioengineered. 2019. PMID: 30876377 Free PMC article. Review.
Cited by
-
Efficient biocatalytic production of D-4-hydroxyphenylglycine by whole cells of recombinant Ralstonia pickettii.Folia Microbiol (Praha). 2009 Nov;54(6):509-15. doi: 10.1007/s12223-009-0073-y. Epub 2010 Feb 7. Folia Microbiol (Praha). 2009. PMID: 20140718
-
Improving the enzymatic activity and stability of N-carbamoyl hydrolase using deep learning approach.Microb Cell Fact. 2024 Jun 4;23(1):164. doi: 10.1186/s12934-024-02439-5. Microb Cell Fact. 2024. PMID: 38834993 Free PMC article.
-
The roles of plant phenolics in defence and communication during Agrobacterium and Rhizobium infection.Mol Plant Pathol. 2010 Sep;11(5):705-19. doi: 10.1111/j.1364-3703.2010.00625.x. Mol Plant Pathol. 2010. PMID: 20696007 Free PMC article. Review.
-
New molecular reporters for rapid protein folding assays.PLoS One. 2008 Jun 11;3(6):e2387. doi: 10.1371/journal.pone.0002387. PLoS One. 2008. PMID: 18545698 Free PMC article.
References
-
- Kleman G. L., Strohl W. R. Developments in high cell density and high productivity microbial fermentation. Curr. Opin. Biotechnol. 1994;5:180–186. - PubMed
-
- Wetzel R. For protein misassembly, it's the ‘‘I’’ decade. Cell. 1996;86:699–702. - PubMed
-
- Goloubinoff P., Gatenby A. A., Lorimer G. H. GroE heat-shock proteins promote assembly of foreign prokaryotic ribulose bisphosphate carboxylase oligomers in Escherichia coli. Nature. 1989;337:44–47. - PubMed
-
- Lee S. C., Olins P. O. Effect of overproduction of heat shock chaperones GroESL and DnaK on human procollagenase production in Escherichia coli. J. Biol. Chem. 1992;267:2849–2852. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

