The Tim9p/10p and Tim8p/13p complexes bind to specific sites on Tim23p during mitochondrial protein import
- PMID: 17122363
- PMCID: PMC1783793
- DOI: 10.1091/mbc.e06-06-0546
The Tim9p/10p and Tim8p/13p complexes bind to specific sites on Tim23p during mitochondrial protein import
Abstract
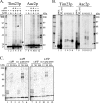
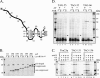
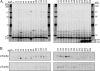
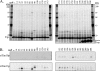
The import of polytopic membrane proteins into the mitochondrial inner membrane (IM) is facilitated by Tim9p/Tim10p and Tim8p/Tim13p protein complexes in the intermembrane space (IMS). These complexes are proposed to act as chaperones by transporting the hydrophobic IM proteins through the aqueous IMS and preventing their aggregation. To examine the nature of this interaction, Tim23p molecules containing a single photoreactive cross-linking probe were imported into mitochondria in the absence of an IM potential where they associated with small Tim complexes in the IMS. On photolysis and immunoprecipitation, a probe located at a particular Tim23p site (27 different locations were examined) was found to react covalently with, in most cases, only one of the small Tim proteins. Tim8p, Tim9p, Tim10p, and Tim13p were therefore positioned adjacent to specific sites in the Tim23p substrate before its integration into the IM. This specificity of binding to Tim23p strongly suggests that small Tim proteins do not function solely as general chaperones by minimizing the exposure of nonpolar Tim23p surfaces to the aqueous medium, but may also align a folded Tim23p substrate in the proper orientation for delivery and integration into the IM at the TIM22 translocon.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

