Dusty protein kinases: primary structure, gene evolution, tissue specific expression and unique features of the catalytic domain
- PMID: 17123648
- PMCID: PMC4277547
- DOI: 10.1016/j.bbaexp.2006.10.004
Dusty protein kinases: primary structure, gene evolution, tissue specific expression and unique features of the catalytic domain
Abstract
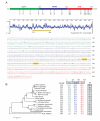
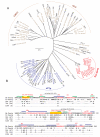
Ser/Thr- and Tyr-Protein kinases constitute a key switch underlying the dynamic nature and graded regulation of signal transduction and pathway activities in cellular organization. Here we describe the identification and characterization of Dusty, a single-copy gene that arose in metazoan evolution and encodes a putative dual Ser/Thr and Tyr protein kinase with unique structural features. Dusty is widely expressed in vertebrates, broadly distributed in the central nervous system, and deregulated in certain human cancers. Confocal imaging of transiently expressed human Dusty-GFP fusion proteins showed a cytoplasmic distribution. Dusty proteins from lower to higher species display an increasing degree of sequence conservation from the N-terminal non-catalytic domain to C-terminal catalytic domain. The non-catalytic region has eight conserved cysteine residues, multiple potential kinase-docking motifs and phosphorylation sites, whereas the catalytic domain is divergent and about equally distant of Ser/Thr and Tyr protein kinases. Homology analyses identified the essential catalytic residues, suggesting that Dusty homologues all possess the enzymatic activity of a protein kinase. Taken together, Dusty is a unique evolutionarily selected group of divergent protein kinases that may play important functional roles in the brain and other tissues of vertebrates.
Figures






References
-
- Kennelly PJ, Krebs EG. Consensus sequences as substrate specificity determinants for protein kinases and protein phosphatases. J. Biol. Chem. 1991;266:15555–15558. - PubMed
-
- Hunter T. Protein kinases and phosphatases: The Yin and Yang of protein phosphorylation and signaling. Cell. 1995;80:2 25–236. - PubMed
-
- Pawson T, Scott JD. Protein phosphorylation in signaling – 50 years and counting. Trends Biochem. Sci. 2005;30:286–290. - PubMed
-
- Hanks SK, Quinn A-M, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. - PubMed
-
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

