Display technologies: application for the discovery of drug and gene delivery agents
- PMID: 17123658
- PMCID: PMC1847402
- DOI: 10.1016/j.addr.2006.09.018
Display technologies: application for the discovery of drug and gene delivery agents
Abstract
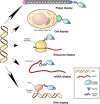
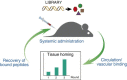
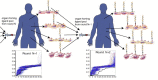
Recognition of molecular diversity of cell surface proteomes in disease is essential for the development of targeted therapies. Progress in targeted therapeutics requires establishing effective approaches for high-throughput identification of agents specific for clinically relevant cell surface markers. Over the past decade, a number of platform strategies have been developed to screen polypeptide libraries for ligands targeting receptors selectively expressed in the context of various cell surface proteomes. Streamlined procedures for identification of ligand-receptor pairs that could serve as targets in disease diagnosis, profiling, imaging and therapy have relied on the display technologies, in which polypeptides with desired binding profiles can be serially selected, in a process called biopanning, based on their physical linkage with the encoding nucleic acid. These technologies include virus/phage display, cell display, ribosomal display, mRNA display and covalent DNA display (CDT), with phage display being by far the most utilized. The scope of this review is the recent advancements in the display technologies with a particular emphasis on molecular mapping of cell surface proteomes with peptide phage display. Prospective applications of targeted compounds derived from display libraries in the discovery of targeted drugs and gene therapy vectors are discussed.
Figures

References
-
- Engwegen J.Y., Gast M.C., Schellens J.H., Beijnen J.H. Clinical proteomics: searching for better tumour markers with SELDI-TOF mass spectrometry. Trends Pharmacol. Sci. 2006;27:251–259. - PubMed
-
- Pasqualini R., Arap W. Vascular targeting. In: Bertino J.R., editor. The Encyclopedia of Cancer. Academic Press; New Brunswick, NJ: 2002. pp. 525–530.
-
- Ruoslahti E. Drug targeting to specific vascular sites. Drug Discov. Today. 2002;7:1138–1143. - PubMed
-
- Molldrem J.J., Komanduri K., Wieder E. Overexpressed differentiation antigens as targets of graft-versus-leukemia reactions. Curr. Opin. Hematol. 2002;9:503–508. - PubMed
-
- Arrell D.K., Neverova I., Van Eyk J.E. Cardiovascular proteomics: evolution and potential. Circ. Res. 2001;88:763–773. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

