Analysis of transcriptional modulation of the presenilin 1 gene promoter by ZNF237, a candidate binding partner of the Ets transcription factor ERM
- PMID: 17126306
- PMCID: PMC1876729
- DOI: 10.1016/j.brainres.2006.10.056
Analysis of transcriptional modulation of the presenilin 1 gene promoter by ZNF237, a candidate binding partner of the Ets transcription factor ERM
Abstract
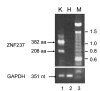
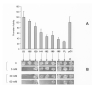
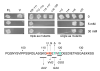
DNA sequences required for the expression of the human presenilin 1 (PS1) gene have been identified between -118 and +178 flanking the major initiation site (+1) mapped in SK-N-SH cells. Several Ets sites are located both upstream as well as downstream from the +1 site, including an Ets motif present at -10 that controls 90% of transcription in SK-N-SH cells. However, in SH-SY5Y cells, transcription initiates further downstream and requires an alternative set of promoter elements including a +90 Ets motif. Ets2, ER81, ERM and Elk1 were identified by yeast one-hybrid selection in a human brain cDNA library using the -10 Ets motif as a bait. We have shown that ERM recognizes specifically Ets motifs on the PS1 promoter located at -10 as well as downstream at +90, +129 and +165 and activates PS1 transcription with promoter fragments whether or not they contain the -10 Ets site. We have now searched for ERM interacting proteins by yeast two-hybrid selection in a human brain cDNA library using the C-terminal 415 amino acid of ERM as a bait. One of the interacting proteins was ZNF237, a member of the MYM gene family. It is widely expressed in different tissues in eukaryotes under several forms derived by alternative splicing, including a large 382 amino acid form containing a single MYM domain, and 2 shorter forms of 208 and 213 amino acids respectively that do not. We show that both the 382 as well as the 208 amino acid forms are expressed in SK-N-SH cells but not in SH-SY5Y cells. Both forms interact with ERM and repress the transcription of PS1 in SH-SY5Y cells. The effect of both C-terminal and N-terminal deletions indicates that the N-terminal 120 amino acid region is required for interaction with ERM in yeast, and furthermore single amino acid mutations show that residues 112 and 114 play an important role. The repression of transcription in SH-SY5Y cells also appears to require the N-terminal potion of ZNF237 and was affected by mutation of the amino acid 112. Data from electrophoretic mobility shift assays indicate that ERM and possibly ZNF237 interact with a fragment of the PS1 promoter.
Figures


5′-AGCAATGCCTCCTGCACCACCAAC-3′
and antisense 5′-CTGCTTCACCACCTTCTTGATG-3′.

5′-AGCAATGCCTCCTGCACCACCAAC-3′
and antisense 5′-CTGCTTCACCACCTTCTTGATG-3′.






Similar articles
-
Alternative initiation of transcription of the human presenilin 1 gene in SH-SY5Y and SK-N-SH cells. The role of Ets factors in the regulation of presenilin 1.Eur J Biochem. 2004 Nov;271(22):4485-94. doi: 10.1111/j.1432-1033.2004.04453.x. Eur J Biochem. 2004. PMID: 15560789
-
The C-terminal region of CHD3/ZFH interacts with the CIDD region of the Ets transcription factor ERM and represses transcription of the human presenilin 1 gene.FEBS J. 2007 Mar;274(6):1434-48. doi: 10.1111/j.1742-4658.2007.05684.x. FEBS J. 2007. PMID: 17489097
-
Ets transcription factors ER81 and Elk1 regulate the transcription of the human presenilin 1 gene promoter.Brain Res Mol Brain Res. 2003 May 12;113(1-2):57-66. doi: 10.1016/s0169-328x(03)00090-1. Brain Res Mol Brain Res. 2003. PMID: 12750007
-
Roles and regulations of the ETS transcription factor ELF4/MEF.J Mol Cell Biol. 2017 Jun 1;9(3):168-177. doi: 10.1093/jmcb/mjw051. J Mol Cell Biol. 2017. PMID: 27932483 Free PMC article. Review.
-
Yeast one-hybrid assays: a historical and technical perspective.Methods. 2012 Aug;57(4):441-7. doi: 10.1016/j.ymeth.2012.07.027. Epub 2012 Aug 3. Methods. 2012. PMID: 22884952 Free PMC article. Review.
Cited by
-
Amyloid Precursor Protein and Alzheimer's Disease.Int J Mol Sci. 2023 Sep 30;24(19):14794. doi: 10.3390/ijms241914794. Int J Mol Sci. 2023. PMID: 37834241 Free PMC article. Review.
-
Ammonia as a Potential Neurotoxic Factor in Alzheimer's Disease.Front Mol Neurosci. 2016 Aug 8;9:57. doi: 10.3389/fnmol.2016.00057. eCollection 2016. Front Mol Neurosci. 2016. PMID: 27551259 Free PMC article. Review.
-
Transcriptional regulation and its misregulation in Alzheimer's disease.Mol Brain. 2013 Oct 21;6:44. doi: 10.1186/1756-6606-6-44. Mol Brain. 2013. PMID: 24144318 Free PMC article. Review.
-
E26 transformation-specific transcription variant 5 in development and cancer: modification, regulation and function.J Biomed Sci. 2023 Mar 6;30(1):17. doi: 10.1186/s12929-023-00909-3. J Biomed Sci. 2023. PMID: 36872348 Free PMC article. Review.
-
Investigation of Glycosylphosphatidylinositol (GPI)-Plasma Membrane Interaction in Live Cells and the Influence of GPI Glycan Structure on the Interaction.Chemistry. 2024 Feb 7;30(8):e202303047. doi: 10.1002/chem.202303047. Epub 2023 Dec 14. Chemistry. 2024. PMID: 37966101 Free PMC article.
References
-
- Baumann H, Kunapuli P, Tracy E, Cowell JK. The oncogenic fusion protein-tyrosine kinase ZNF198/fibroblast growth factor receptor-1 has signaling function comparable with interleukin-6 cytokine receptors. J Biol Chem. 2003;278:16198–16208. - PubMed
-
- Chen J, Deangelo DJ, Kutok JL, Williams IR, Lee BH, Wadleigh M, Duclos N, Cohen S, Adelsperger J, Okabe R, Coburn A, Galinsky I, Huntly B, Cohen PS, Meyer T, Fabbro D, Roesel J, Banerji L, Griffin JD, Xiao S, Fletcher JA, Stone RM, Gilliland DG. PKC412 inhibits the zinc finger 198-fibroblast growth factor receptor 1 fusion tyrosine kinase and is active in treatment of stem cell myeloproliferative disorder. Proc Natl Acad Sci U S A. 2004;101:14479–14484. - PMC - PubMed
-
- Chotteau-Lelievre A, Dolle P, Peronne V, Coutte L, de Launoit Y, Desbiens X. Expression patterns of the Ets transcription factors from the PEA3 group during early stages of mouse development. Mech Dev. 2001;108:191–195. - PubMed
-
- Chotteau-Lelievre A, Montesano R, Soriano J, Soulie P, Desbiens X, de Launoit Y. PEA3 transcription factors are expressed in tissues undergoing branching morphogenesis and promote formation of duct-like structures by mammary epithelial cells in vitro. Dev Biol. 2003;259:241–257. - PubMed
-
- Chyung JH, Raper DM, Selkoe DJ. Gamma-secretase exists on the plasma membrane as an intact complex that accepts substrates and effects intramembrane cleavage. J Biol Chem. 2005;280:4383–4392. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

