Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
- PMID: 17129781
- PMCID: PMC1876690
- DOI: 10.1016/j.cell.2006.11.023
Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Abstract
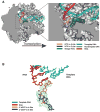
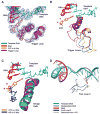
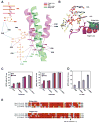
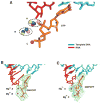
New structures of RNA polymerase II (pol II) transcribing complexes reveal a likely key to transcription. The trigger loop swings beneath a correct nucleoside triphosphate (NTP) in the nucleotide addition site, closing off the active center and forming an extensive network of interactions with the NTP base, sugar, phosphates, and additional pol II residues. A histidine side chain in the trigger loop, precisely positioned by these interactions, may literally "trigger" phosphodiester bond formation. Recognition and catalysis are thus coupled, ensuring the fidelity of transcription.
Figures

References
-
- Albert DA, Gudas LJ. Ribonucleotide reductase activity and deoxyribonucleoside triphosphate metabolism during the cell cycle of S49 wild-type and mutant mouse T-lymphoma cells. J Biol Chem. 1985;260:679–684. - PubMed
-
- Allison LA, Moyle M, Shales M, Ingles CJ. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985;42:599–610. - PubMed
-
- Artsimovitch I, Patlan V, Sekine S, Vassylyeva MN, Hosaka T, Ochi K, Yokoyama S, Vassylyev DG. Structural basis for transcription regulation by alarmone ppGpp. Cell. 2004;117:299–310. - PubMed
-
- Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–193. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

