ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis
- PMID: 17130150
- PMCID: PMC1716726
- DOI: 10.1093/nar/gkl783
ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis
Abstract
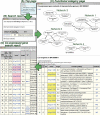
Publicly available database of co-expressed gene sets would be a valuable tool for a wide variety of experimental designs, including targeting of genes for functional identification or for regulatory investigation. Here, we report the construction of an Arabidopsis thaliana trans-factor and cis-element prediction database (ATTED-II) that provides co-regulated gene relationships based on co-expressed genes deduced from microarray data and the predicted cis elements. ATTED-II (http://www.atted.bio.titech.ac.jp) includes the following features: (i) lists and networks of co-expressed genes calculated from 58 publicly available experimental series, which are composed of 1388 GeneChip data in A.thaliana; (ii) prediction of cis-regulatory elements in the 200 bp region upstream of the transcription start site to predict co-regulated genes amongst the co-expressed genes; and (iii) visual representation of expression patterns for individual genes. ATTED-II can thus help researchers to clarify the function and regulation of particular genes and gene networks.
Figures

Similar articles
-
AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors.BMC Bioinformatics. 2003 Jun 23;4:25. doi: 10.1186/1471-2105-4-25. Epub 2003 Jun 23. BMC Bioinformatics. 2003. PMID: 12820902 Free PMC article.
-
POXO: a web-enabled tool series to discover transcription factor binding sites.Nucleic Acids Res. 2006 Jul 1;34(Web Server issue):W534-40. doi: 10.1093/nar/gkl296. Nucleic Acids Res. 2006. PMID: 16845065 Free PMC article.
-
AthaMap web tools for the analysis and identification of co-regulated genes.Nucleic Acids Res. 2007 Jan;35(Database issue):D857-62. doi: 10.1093/nar/gkl1006. Epub 2006 Dec 5. Nucleic Acids Res. 2007. PMID: 17148485 Free PMC article.
-
Approaches for extracting practical information from gene co-expression networks in plant biology.Plant Cell Physiol. 2007 Mar;48(3):381-90. doi: 10.1093/pcp/pcm013. Epub 2007 Jan 23. Plant Cell Physiol. 2007. PMID: 17251202 Review.
-
trans meets cis in MADS science.Trends Plant Sci. 2006 May;11(5):224-31. doi: 10.1016/j.tplants.2006.03.008. Epub 2006 Apr 17. Trends Plant Sci. 2006. PMID: 16616581 Review.
Cited by
-
Arabidopsis ensemble reverse-engineered gene regulatory network discloses interconnected transcription factors in oxidative stress.Plant Cell. 2014 Dec;26(12):4656-79. doi: 10.1105/tpc.114.131417. Epub 2014 Dec 30. Plant Cell. 2014. PMID: 25549671 Free PMC article.
-
Coexpression landscape in ATTED-II: usage of gene list and gene network for various types of pathways.J Plant Res. 2010 May;123(3):311-9. doi: 10.1007/s10265-010-0333-6. Epub 2010 Apr 10. J Plant Res. 2010. PMID: 20383554 Review.
-
RNA-Seq analysis of differential gene expression in Betula luminifera xylem during the early stages of tension wood formation.PeerJ. 2018 Aug 21;6:e5427. doi: 10.7717/peerj.5427. eCollection 2018. PeerJ. 2018. PMID: 30155351 Free PMC article.
-
The sugar transporter SWEET10 acts downstream of FLOWERING LOCUS T during floral transition of Arabidopsis thaliana.BMC Plant Biol. 2020 Feb 3;20(1):53. doi: 10.1186/s12870-020-2266-0. BMC Plant Biol. 2020. PMID: 32013867 Free PMC article.
-
Establishment of an in planta magnesium monitoring system using CAX3 promoter-luciferase in Arabidopsis.J Exp Bot. 2012 Jan;63(1):355-63. doi: 10.1093/jxb/err283. Epub 2011 Sep 13. J Exp Bot. 2012. PMID: 21914662 Free PMC article.
References
-
- Ikeo K., Ishi-i J., Tamura T., Gojobori T., Tateno Y. CIBEX: center for information biology gene expression database. C. R. Biol. 2003;326:1079–1082. - PubMed
-
- Rhee S.Y., Beavis W., Berardini T.Z., Chen G., Dixon D., Doyle A., Garcia-Hernandez M., Huala E., Lander G., Montoya M., et al. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003;31:224–228. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

