Self-splicing of a group IIC intron: 5' exon recognition and alternative 5' splicing events implicate the stem-loop motif of a transcriptional terminator
- PMID: 17130159
- PMCID: PMC1702495
- DOI: 10.1093/nar/gkl820
Self-splicing of a group IIC intron: 5' exon recognition and alternative 5' splicing events implicate the stem-loop motif of a transcriptional terminator
Abstract
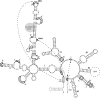
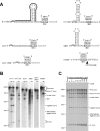
Bacterial IIC introns are a newly recognized subclass of group II introns whose ribozyme properties have not been characterized in detail. IIC introns are typically located downstream of transcriptional terminator motifs (inverted repeat followed by T's) or other inverted repeats in bacterial genomes. Here we have characterized the self-splicing activity of a IIC intron, B.h.I1, from Bacillus halodurans. B.h.I1 self-splices in vitro through hydrolysis to produce linear intron, but interestingly, additional unexpected products were formed that were highly dependent on ionic conditions. These products were determined to represent alternative splicing events at the 5' junction and cleavages throughout the RNA transcript. The alternative splicing and cleavage events occurred at cryptic splice sites containing stem-loop and IBS1 motifs, suggesting that the 5' exon is recognized by both elements. These results provide the first example of a group II intron that uses 5' splice sites nonadjacent to the ribozyme structure. Furthermore, the data suggest that IIC introns differ from IIA and IIB introns with respect to 5' exon definition, and that the terminator stem-loop substitutes in part for the missing IBS2-EBS2 (intron and exon binding sites 2) interaction.
Figures


References
-
- Robart A.R., Zimmerly S. Group II intron retroelements: function and diversity. Cytogenet. Genome Res. 2005;110:589–597. - PubMed
-
- Lambowitz A.M., Zimmerly S. Mobile group II introns. Annu. Rev. Genet. 2004;38:1–35. - PubMed
-
- Toro N. Bacteria and archaea group II introns: additional mobile genetic elements in the environment. Environ. Microbiol. 2003;5:143–151. - PubMed
-
- Belfort M., Derbyshire V., Parker M.M., Cousineau B., Lambowitz A.M. Mobile introns: pathways and proteins. In: Craig N.L., Craigie R., Gellert M., Lambowitz A.M., editors. Mobile DNA II. Washington DC: ASM Press; 2002. pp. 761–783.
-
- Bonen L., Vogel J. The ins and outs of group II introns. Trends Genet. 2001;17:322–331. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

