Molecular dysfunction associated with the human mitochondrial 3302A>G mutation in the MTTL1 (mt-tRNALeu(UUR)) gene
- PMID: 17130166
- PMCID: PMC1702489
- DOI: 10.1093/nar/gkl727
Molecular dysfunction associated with the human mitochondrial 3302A>G mutation in the MTTL1 (mt-tRNALeu(UUR)) gene
Abstract
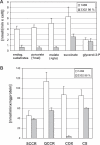
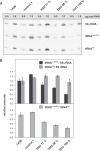
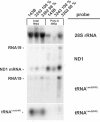
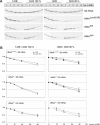
The gene encoding mt-tRNA(Leu(UUR)), MT-TL1, is a hotspot for pathogenic mtDNA mutations. Amongst the first to be described was the 3302A>G transition which resulted in a substantial accumulation in patient muscle of RNA19, an unprocessed RNA intermediate including mt-16S rRNA, mt-tRNA(Leu(UUR)) and MTND1. We have now been able to further assess the molecular aetiology associated with 3302A>G in transmitochondrial cybrids. Increased steady-state levels of RNA19 was confirmed, although not to the levels previously reported in muscle. This data was consistent with an increase in RNA19 stability. The mutation resulted in decreased mt-tRNA(Leu(UUR)) levels, but its stability was unchanged, consistent with a defect in RNA19 processing responsible for low tRNA levels. A partial defect in aminoacylation was also identified, potentially caused by an alteration in tRNA structure. These deficiencies lead to a severe defect in respiration in the transmitochondrial cybrids, consistent with the profound mitochondrial disorder originally associated with this mutation.
Figures




Similar articles
-
The pathogenic A3243G mutation in human mitochondrial tRNALeu(UUR) decreases the efficiency of aminoacylation.Biochemistry. 2003 Feb 4;42(4):958-64. doi: 10.1021/bi026882r. Biochemistry. 2003. PMID: 12549915
-
Mitochondrial tRNA genes are hotspots for mutations in a cohort of patients with exercise intolerance and mitochondrial myopathy.J Neurol Sci. 2017 Aug 15;379:137-143. doi: 10.1016/j.jns.2017.05.056. Epub 2017 May 30. J Neurol Sci. 2017. PMID: 28716227
-
Correction of the consequences of mitochondrial 3243A>G mutation in the MT-TL1 gene causing the MELAS syndrome by tRNA import into mitochondria.Nucleic Acids Res. 2011 Oct;39(18):8173-86. doi: 10.1093/nar/gkr546. Epub 2011 Jun 30. Nucleic Acids Res. 2011. PMID: 21724600 Free PMC article.
-
The molecular pathology of pathogenic mitochondrial tRNA variants.FEBS Lett. 2021 Apr;595(8):1003-1024. doi: 10.1002/1873-3468.14049. Epub 2021 Feb 12. FEBS Lett. 2021. PMID: 33513266 Free PMC article. Review.
-
The non-syndromic clinical spectrums of mtDNA 3243A>G mutation.Neurosciences (Riyadh). 2021 Apr;26(2):128-133. doi: 10.17712/nsj.2021.2.20200145. Neurosciences (Riyadh). 2021. PMID: 33814365 Free PMC article. Review.
Cited by
-
Human mtDNA-Encoded Long ncRNAs: Knotty Molecules and Complex Functions.Int J Mol Sci. 2024 Jan 25;25(3):1502. doi: 10.3390/ijms25031502. Int J Mol Sci. 2024. PMID: 38338781 Free PMC article. Review.
-
Human mitochondrial transcription revisited: only TFAM and TFB2M are required for transcription of the mitochondrial genes in vitro.J Biol Chem. 2010 Jun 11;285(24):18129-33. doi: 10.1074/jbc.C110.128918. Epub 2010 Apr 21. J Biol Chem. 2010. PMID: 20410300 Free PMC article.
-
High glucose uptake unexpectedly is accompanied by high levels of the mitochondrial ß-F1-ATPase subunit in head and neck squamous cell carcinoma.Oncotarget. 2015 Nov 3;6(34):36172-84. doi: 10.18632/oncotarget.5459. Oncotarget. 2015. PMID: 26452026 Free PMC article.
-
Simultaneous DNA and RNA Mapping of Somatic Mitochondrial Mutations across Diverse Human Cancers.PLoS Genet. 2015 Jun 30;11(6):e1005333. doi: 10.1371/journal.pgen.1005333. eCollection 2015 Jun. PLoS Genet. 2015. PMID: 26125550 Free PMC article.
-
Myoclonic epilepsy with ragged red fibers syndrome associated with mitochondrial 3302A>G mutation in the MT‑TL1 gene: A case report.Exp Ther Med. 2023 Jan 4;25(2):87. doi: 10.3892/etm.2023.11786. eCollection 2023 Feb. Exp Ther Med. 2023. PMID: 36684660 Free PMC article.
References
-
- Bindoff L.A., Howell N., Poulton J., McCullough D.A., Morten K.J., Lightowlers R.N., Turnbull D.M., Weber K. Abnormal RNA processing associated with a novel tRNA mutation in mitochondrial DNA. A potential disease mechanism. J. Biol. Chem. 1993;268:19559–19564. - PubMed
-
- Van Den Bosch B.J., De Coo I.F., Hendrickx A.T., Busch H.F., De Jong G., Scholte H.R., Smeets H.J. Increased risk for cardiorespiratory failure associated with the A3302G mutation in the mitochondrial DNA encoded tRNA(Leu(UUR)) gene. Neuromuscul. Disord. 2004;14:683–688. - PubMed
-
- King M.P., Attardi G. Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science. 1989;246:500–503. - PubMed

