Genome-wide loss of heterozygosity and copy number alteration in esophageal squamous cell carcinoma using the Affymetrix GeneChip Mapping 10 K array
- PMID: 17134496
- PMCID: PMC1687196
- DOI: 10.1186/1471-2164-7-299
Genome-wide loss of heterozygosity and copy number alteration in esophageal squamous cell carcinoma using the Affymetrix GeneChip Mapping 10 K array
Abstract
Background: Esophageal squamous cell carcinoma (ESCC) is a common malignancy worldwide. Comprehensive genomic characterization of ESCC will further our understanding of the carcinogenesis process in this disease.
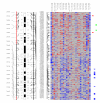
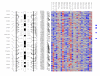
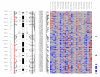
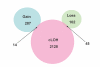
Results: Genome-wide detection of chromosomal changes was performed using the Affymetrix GeneChip 10 K single nucleotide polymorphism (SNP) array, including loss of heterozygosity (LOH) and copy number alterations (CNA), for 26 pairs of matched germ-line and micro-dissected tumor DNA samples. LOH regions were identified by two methods--using Affymetrix's genotype call software and using Affymetrix's copy number alteration tool (CNAT) software--and both approaches yielded similar results. Non-random LOH regions were found on 10 chromosomal arms (in decreasing order of frequency: 17p, 9p, 9q, 13q, 17q, 4q, 4p, 3p, 15q, and 5q), including 20 novel LOH regions (10 kb to 4.26 Mb). Fifteen CNA-loss regions (200 kb to 4.3 Mb) and 36 CNA-gain regions (200 kb to 9.3 Mb) were also identified.
Conclusion: These studies demonstrate that the Affymetrix 10 K SNP chip is a valid platform to integrate analyses of LOH and CNA. The comprehensive knowledge gained from this analysis will enable improved strategies to prevent, diagnose, and treat ESCC.
Figures



References
-
- Phillips MS, Lawrence R, Sachidanandam R, Morris AP, Balding DJ, Donaldson MA, Studebaker JF, Ankener WM, Alfisi SV, Kuo FS, Camisa AL, Pazorov V, Scott KE, Carey BJ, Faith J, Katari G, Bhatti HA, Cyr JM, Derohannessian V, Elosua C, Forman AM, Grecco NM, Hock CR, Kuebler JM, Lathrop JA, Mockler MA, Nachtman EP, Restine SL, Varde SA, Hozza MJ, Gelfand CA, Broxholme J, Abecasis GR, Boyce-Jacino MT, Cardon LR. Chromosome-wide distribution of haplotype blocks and the role of recombination hot spots. Nat Genet. 2003;33:382–387. doi: 10.1038/ng1100. - DOI - PubMed
-
- Zhao X, Li C, Paez JG, Chin K, Janne PA, Chen TH, Girard L, Minna J, Christiani D, Leo C, Gray JW, Sellers WR, Meyerson M. An integrated view of copy number and allelic alterations in the cancer genome using single nucleotide polymorphism arrays. Cancer Res. 2004;64:3060–3071. doi: 10.1158/0008-5472.CAN-03-3308. - DOI - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

