Effect of infliximab on mRNA expression profiles in synovial tissue of rheumatoid arthritis patients
- PMID: 17134501
- PMCID: PMC1794525
- DOI: 10.1186/ar2090
Effect of infliximab on mRNA expression profiles in synovial tissue of rheumatoid arthritis patients
Abstract
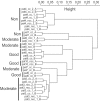
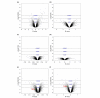
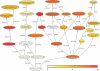
We examined the gene expression profiles in arthroscopic biopsies retrieved from 10 rheumatoid arthritis patients before and after anti-TNF treatment with infliximab to investigate whether such profiles can be used to predict responses to the therapy, and to study effects of the therapy on the profiles. Responses to treatment were assessed using European League Against Rheumatism response criteria. Three patients were found to be good responders, five patients to be moderate responders and two patients to be nonresponders. The TNF-alpha status of the biopsies from each of the patients before treatment was also investigated immunohistochemically, and it was detected in biopsies from four of the patients, including all three of the good responders. The gene expression data demonstrate that all patients had unique gene expression signatures, with low intrapatient variability between biopsies. The data also revealed significant differences between the good responding and nonresponding patients (279 differentially expressed genes were detected, with a false discovery rate < 0.025). Among the identified genes we found that MMP-3 was significantly upregulated in good responders (log2 fold change, 2.95) compared with nonresponders, providing further support for the potential of MMP-3 as a marker for good responses to therapy. An even more extensive list of 685 significantly differentially expressed genes was found between patients in whom TNF-alpha was found and nonresponders, indicating that TNF-alpha could be an important biomarker for successful infliximab treatment. Significant differences were also observed between biopsies taken before and after anti-TNF treatment, including 115 differentially expressed genes in the good responding group. Interestingly, the effect was even stronger in the group in which TNF-alpha was immunohistochemically detected before therapy. Here, 1,058 genes were differentially expressed, including many that were novel in this context (for example, CXCL3 and CXCL14). Subsequent Gene Ontology analysis revealed that several 'themes' were significantly over-represented that are known to be affected by anti-TNF treatment in inflammatory tissue; for example, immune response (GO:0006955), cell communication (GO:0007154), signal transduction (GO:0007165) and chemotaxis (GO:0006935). No genes reached statistical significance in the moderately responding or nonresponding groups. In conclusion, this pilot study suggests that further investigation is warranted on the usefulness of gene expression profiling of synovial tissue to predict and monitor the outcome of rheumatoid arthritis therapies.
Figures
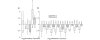
References
-
- Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, Healey LA, Kaplan SR, Liang MH, Luthra HS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis Rheum. 1988;31:315–324. - PubMed
-
- van der Pouw Kraan TC, van Gaalen FA, Kasperkovitz PV, Verbeet NL, Smeets TJ, Kraan MC, Fero M, Tak PP, Huizinga TW, Pieterman E, et al. Rheumatoid arthritis is a heterogeneous disease: evidence for differences in the activation of the STAT-1 pathway between rheumatoid tissues. Arthritis Rheum. 2003;48:2132–2145. doi: 10.1002/art.11096. - DOI - PubMed
-
- van der Pouw Kraan TC, van Gaalen FA, Huizinga TW, Pieterman E, Breedveld FC, Verweij CL. Discovery of distinctive gene expression profiles in rheumatoid synovium using cDNA microarray technology: evidence for the existence of multiple pathways of tissue destruction and repair. Genes Immun. 2003;4:187–196. doi: 10.1038/sj.gene.6363975. - DOI - PubMed
-
- Maini R, St Clair EW, Breedveld F, Furst D, Kalden J, Weisman M, Smolen J, Emery P, Harriman G, Feldmann M, et al. Infliximab (chimeric anti-tumour necrosis factor alpha monoclonal antibody) versus placebo in rheumatoid arthritis patients receiving concomitant methotrexate: a randomised phase III trial. ATTRACT Study Group. Lancet. 1999;354:1932–1939. doi: 10.1016/S0140-6736(99)05246-0. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

