MBGD: a platform for microbial comparative genomics based on the automated construction of orthologous groups
- PMID: 17135196
- PMCID: PMC1751538
- DOI: 10.1093/nar/gkl978
MBGD: a platform for microbial comparative genomics based on the automated construction of orthologous groups
Abstract
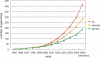
The microbial genome database for comparative analysis (MBGD) is a comprehensive platform for microbial comparative genomics. The central function of MBGD is to create orthologous groups among multiple genomes from precomputed all-against-all similarity relationships using the DomClust algorithm. The database now contains >300 published genomes and the number continues to grow. For researchers who are interested in ongoing genome projects, we have now started a new service called 'My MBGD,' which allows users to add their own genome sequences to MBGD for the purpose of identifying orthologs among both the new and the existing genomes. Furthermore, in order to make available the rapidly accumulating information on closely related genome sequences, we enhanced the interface for pairwise genome comparisons using the CGAT interface, which allows users to see nucleotide sequence alignments of non-coding as well as coding regions. MBGD is available at http://mbgd.genome.ad.jp/.
Figures