Tomato chlorotic mottle virus is a target of RNA silencing but the presence of specific short interfering RNAs does not guarantee resistance in transgenic plants
- PMID: 17135316
- PMCID: PMC1797551
- DOI: 10.1128/JVI.01238-06
Tomato chlorotic mottle virus is a target of RNA silencing but the presence of specific short interfering RNAs does not guarantee resistance in transgenic plants
Abstract
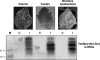
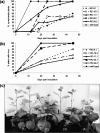
Tomato chlorotic mottle virus (ToCMoV) is a begomovirus found widespread in tomato fields in Brazil. ToCMoV isolate BA-Se1 (ToCMoV-[BA-Se1]) was shown to trigger the plant RNA silencing surveillance in different host plants and, coinciding with a decrease in viral DNA levels, small interfering RNAs (siRNAs) specific to ToCMoV-[BA-Se1] accumulated in infected plants. Although not homogeneously distributed, the siRNA population in both infected Nicotiana benthamiana and tomato plants represented the entire DNA-A and DNA-B genomes. We determined that in N. benthamiana, the primary targets corresponded to the 5' end of AC1 and the embedded AC4, the intergenic region and 5' end of AV1 and overlapping central part of AC5. Subsequently, transgenic N. benthamiana plants were generated that were preprogrammed to express double-stranded RNA corresponding to this most targeted portion of the virus genome by using an intron-hairpin construct. These plants were shown to indeed produce ToCMoV-specific siRNAs. When challenge inoculated, most transgenic lines showed significant delays in symptom development, and two lines had immune plants. Interestingly, the levels of transgene-produced siRNAs were similar in resistant and susceptible siblings of the same line. This indicates that, in contrast to RNA viruses, the mere presence of transgene siRNAs corresponding to DNA virus sequences does not guarantee virus resistance and that other factors may play a role in determining RNA-mediated resistance to DNA viruses.
Figures




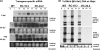
References
-
- Al-Kaff, N. S., S. N. Covey, M. M. Kreike, A. M. Page, R. Pinder, and P. J. Dale. 1998. Transcriptional and posttranscriptional plant gene silencing in response to a pathogen. Science 279:2113-2115. - PubMed
-
- Aragão, F. J. L., S. G. Ribeiro, L. M. G. Barros, A. C. M. Brasileiro, D. P. Maxwell, E. L. Rech, and J. C. Faria. 1998. Transgenic beans (Phaseolus vulgaris L.) engineered to express viral antisense RNAs show delayed and attenuated symptoms to bean golden mosaic geminivirus. Mol. Breed. 4:491-499.
-
- Asad, S., W. A. Haris, A. Bashir, Y. Zafar, K. A. Malik, N. N. Malik, and C. P. Lichtenstein. 2003. Transgenic tobacco expressing geminiviral RNAs are resistant to the serious viral pathogen causing cotton leaf curl disease. Arch. Virol. 148:2341-2352. - PubMed
-
- Azzam, O., J. Frazer, D. De La Rosa, J. S. Beaver, P. Ahlquist, and D. P. Maxwell. 1994. Whitefly transmission and efficient ssDNA accumulation of Bean golden mosaic geminivirus require functional coat protein. Virology 204:289-296. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

