Isolation and characterization of a myo-inositol-1-phosphate synthase gene from yellow passion fruit (Passiflora edulis f. flavicarpa) expressed during seed development and environmental stress
- PMID: 17138579
- PMCID: PMC2802995
- DOI: 10.1093/aob/mcl256
Isolation and characterization of a myo-inositol-1-phosphate synthase gene from yellow passion fruit (Passiflora edulis f. flavicarpa) expressed during seed development and environmental stress
Abstract
Background and aims: Myo-inositol-1l-phosphate synthase (MIPS) catalyses the conversion of d-glucose 6-phosphate to 1-l-myo-inositol-1-phosphate, the first and rate-limiting step in the biosynthesis of all inositol-containing compounds. Inositol phospholipids play a vital role in membrane trafficking and signalling pathways, auxin storage and transport, phytic acid biosynthesis, cell wall biosynthesis and production of stress-related molecules. In the present study, an MIPS cDNA from developing Passiflora edulis f. flavicarpa seeds was characterized and an investigation made into its spatial and differential expression, as well as changes in its transcription during exposure of growing plants to cold and heat stresses.
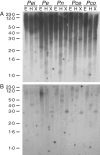
Methods: The MIPS-encoding gene was isolated by polymerase chain reaction (PCR) methods, and transcript levels were examined using semi-quantitative reverse transcription-PCR (RT-PCR) during seed development and in response to heat and cold stress. In addition, the copy number of the cloned PeMIPS1 gene in the genome of Passiflora edulis, P. eichleriana, P. caerulea, P. nitida and P. coccinea was determined by Southern blot analyses.
Key results: A full-length cDNA clone of the PeMIPS1 from P. edulis was isolated and characterized. Southern blot analyses indicated that the genomic DNA might have diverse sequences of MIPS-encoding genes and one copy of the cloned PeMIPS1 gene in the genomes of P. edulis, P. eichleriana, P. caerulea, P. nitida and P. coccinea. RT-PCR expression analyses revealed the presence of PeMIPS1 transcripts in ovules, pollen grains and leaves, and during the seed developmental stages, where it peaked at 9 d after pollination. The PeMIPS1 gene is differentially regulated under cold and heat stress, presenting a light-responsive transcription.
Conclusions: Experimental data suggest that PeMIPS1 transcription plays an important role in the establishment of developmental programmes and during the response of plants to environmental changes. The PeMIPS1 is differentially transcribed during cold and heat stress, presenting a light response pattern, suggesting that it is important for environmental stress response.
Figures



References
-
- Bohnert HJ, Sheveleva E. Plant stress adaptations – making metabolism move. Current Opinion in Plant Biology. 1998;1:267–274. - PubMed
-
- Bray EA, Bailey-Serres J, Weretilnyk E. Response to abiotic stress. In: Buchanan BB, Gruissem W, Jones RL, editors. Biochemistry and molecular biology of plants. Rockville, MD: American Society of Plant Physiologists; 2000. pp. 1158–1203.
-
- Chun JA, Jin UH, Lee JW, Yi YB, Hyung NI, Kang MH. Isolation and characterization of a myo-inositol 1-phosphate synthase cDNA from developing sesame (Sesamum indicum L.) seeds: functional and differential expression, and salt-induced transcription during germination. Planta. 2003;216:874–880. - PubMed
-
- Das-Chatterjee A, Goswami L, Maitra S, Dastidar KG, Ray S, Majumder AL. Introgression of a novel salt-tolerant l-myo-inositol 1-phosphate synthase from Porteresia coarctata (Roxb.) Tateoka (PcINO1) confers salt tolerance to evolutionary diverse organisms. FEBS Letters. 2006;580:3980–3988. - PubMed
MeSH terms
Substances
Associated data
- Actions

