Repression of ADH1 and ADH3 during zinc deficiency by Zap1-induced intergenic RNA transcripts
- PMID: 17139254
- PMCID: PMC1698899
- DOI: 10.1038/sj.emboj.7601453
Repression of ADH1 and ADH3 during zinc deficiency by Zap1-induced intergenic RNA transcripts
Abstract
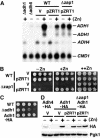
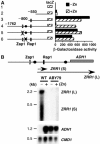
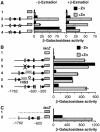
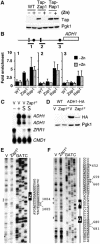
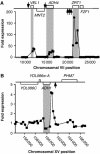
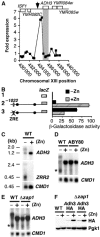
The transcriptional activator Zap1 induces target gene expression in response to zinc deficiency. We demonstrate that during zinc starvation, Zap1 is required for the repression of ADH1 expression. ADH1 encodes the major zinc-dependent alcohol dehydrogenase that is utilized during fermentation. During zinc starvation, Zap1 binds upstream of the activator Rap1 and induces an intergenic RNA transcript, ZRR1. ZRR1 expression leads to the transient displacement of Rap1 from the ADH1 promoter resulting in ADH1 repression. Using a microarray-based approach, we screened for additional genes repressed by Zap1 intergenic transcripts. We found that ADH3, the major mitochondrial alcohol dehydrogenase, is regulated in a manner similar to ADH1. Thus, during zinc deficiency, Zap1 mediates the repression of two of the most abundant zinc-requiring enzymes.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

