Structure of the origin-binding domain of simian virus 40 large T antigen bound to DNA
- PMID: 17139255
- PMCID: PMC1698898
- DOI: 10.1038/sj.emboj.7601452
Structure of the origin-binding domain of simian virus 40 large T antigen bound to DNA
Abstract
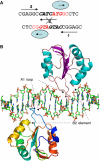
The large T antigen (T-ag) protein binds to and activates DNA replication from the origin of DNA replication (ori) in simian virus 40 (SV40). Here, we determined the crystal structures of the T-ag origin-binding domain (OBD) in apo form, and bound to either a 17 bp palindrome (sites 1 and 3) or a 23 bp ori DNA palindrome comprising all four GAGGC binding sites for OBD. The T-ag OBDs were shown to interact with the DNA through a loop comprising Ser147-Thr155 (A1 loop), a combination of a DNA-binding helix and loop (His203-Asn210), and Asn227. The A1 loop traveled back-and-forth along the major groove and accounted for most of the sequence-determining contacts with the DNA. Unexpectedly, in both T-ag-DNA structures, the T-ag OBDs bound DNA independently and did not make direct protein-protein contacts. The T-ag OBD was also captured bound to a non-consensus site ATGGC even in the presence of its canonical site GAGGC. Our observations taken together with the known biochemical and structural features of the T-ag-origin interaction suggest a model for origin unwinding.
Figures



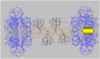

References
-
- Bochkarev A, Barwell JA, Pfuetzner RA, Bochkareva E, Frappier L, Edwards AM (1996) Crystal structure of the DNA-binding domain of the Epstein–Barr virus origin-binding protein, EBNA1, bound to DNA. Cell 84: 791–800 - PubMed
-
- Bochkarev A, Barwell JA, Pfuetzner RA, Furey W Jr, Edwards AM, Frappier L (1995) Crystal structure of the DNA-binding domain of the Epstein–Barr virus origin-binding protein EBNA 1. Cell 83: 39–46 - PubMed
-
- Bullock PA (1997) The initiation of simian virus 40 DNA replication in vitro. Crit Rev Biochem Mol Biol 32: 503–568 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

